Gene Page: KIFC3
Summary ?
| GeneID | 3801 |
| Symbol | KIFC3 |
| Synonyms | - |
| Description | kinesin family member C3 |
| Reference | MIM:604535|HGNC:HGNC:6326|Ensembl:ENSG00000140859|HPRD:05171|Vega:OTTHUMG00000133455 |
| Gene type | protein-coding |
| Map location | 16q13-q21 |
| Pascal p-value | 0.613 |
| Sherlock p-value | 0.363 |
| Fetal beta | -0.511 |
| DMG | 3 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01115923 | 16 | 57793728 | KIFC3 | 3.16E-5 | -0.005 | 0.076 | DMG:Montano_2016 |
| cg07299078 | 16 | 57817313 | KIFC3 | 3.99E-4 | 0.32 | 0.044 | DMG:Wockner_2014 |
| cg07685869 | 16 | 57836706 | KIFC3 | 9.41E-8 | -0.036 | 2.13E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16966868 | chr16 | 57183074 | KIFC3 | 3801 | 0.16 | cis | ||
| rs16827037 | chr3 | 117203284 | KIFC3 | 3801 | 0.07 | trans |
Section II. Transcriptome annotation
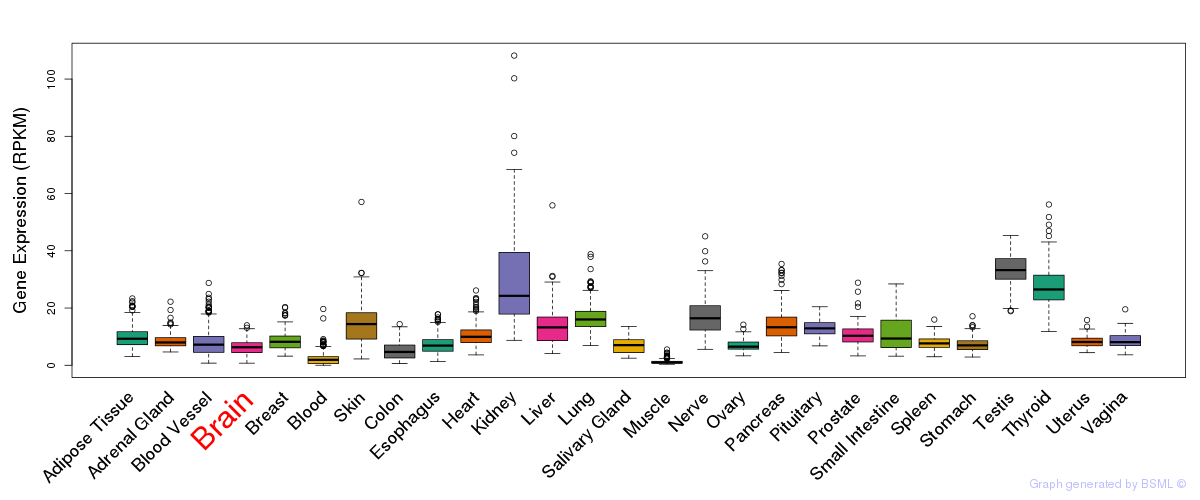
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID ECADHERIN STABILIZATION PATHWAY | 42 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | 27 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME PROTEIN FOLDING | 53 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL UP | 285 | 181 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS UP | 137 | 94 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| ROYLANCE BREAST CANCER 16Q COPY NUMBER UP | 63 | 44 | All SZGR 2.0 genes in this pathway |
| AMUNDSON RESPONSE TO ARSENITE | 217 | 143 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| SUNG METASTASIS STROMA DN | 54 | 34 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 1 UP | 182 | 119 | All SZGR 2.0 genes in this pathway |
| DELASERNA MYOD TARGETS UP | 89 | 51 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE DN | 261 | 183 | All SZGR 2.0 genes in this pathway |
| SAFFORD T LYMPHOCYTE ANERGY | 87 | 54 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED UP | 85 | 50 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 | 227 | 149 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF INTERVAL UP | 105 | 46 | All SZGR 2.0 genes in this pathway |