Gene Page: ARF5
Summary ?
| GeneID | 381 |
| Symbol | ARF5 |
| Synonyms | - |
| Description | ADP ribosylation factor 5 |
| Reference | MIM:103188|HGNC:HGNC:658|Ensembl:ENSG00000004059|HPRD:00055|Vega:OTTHUMG00000023246 |
| Gene type | protein-coding |
| Map location | 7q31.3 |
| Pascal p-value | 0.091 |
| Sherlock p-value | 0.826 |
| Fetal beta | -0.499 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14938677 | 7 | 127231698 | ARF5 | 2.961E-4 | 0.34 | 0.039 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17795337 | chr6 | 136492658 | ARF5 | 381 | 0.06 | trans |
Section II. Transcriptome annotation
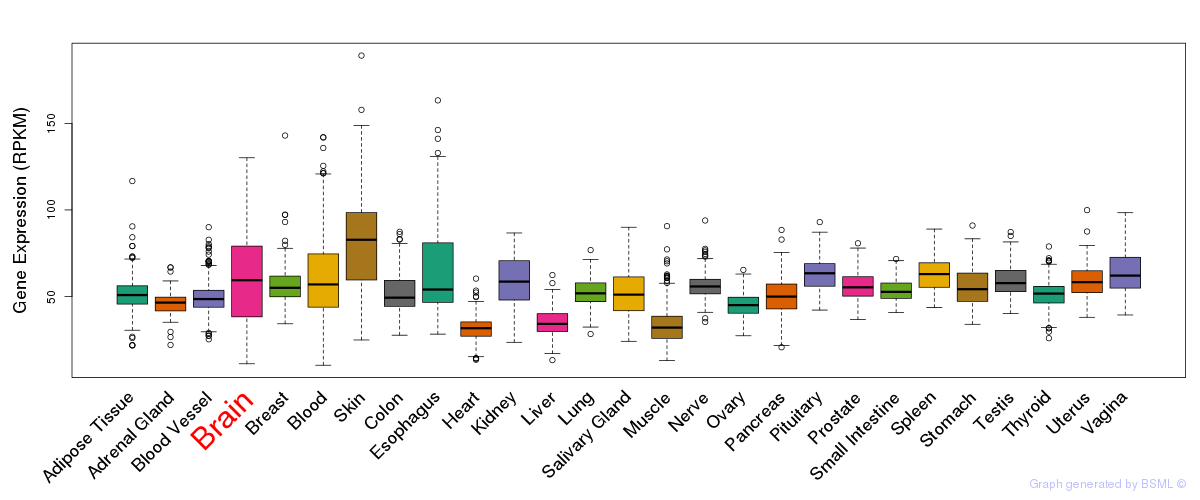
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KARS | 0.93 | 0.92 |
| ACBD6 | 0.92 | 0.92 |
| NIPSNAP1 | 0.92 | 0.94 |
| IGBP1 | 0.91 | 0.92 |
| CDK8 | 0.91 | 0.92 |
| CCT3 | 0.91 | 0.93 |
| RFWD2 | 0.91 | 0.94 |
| PTDSS1 | 0.90 | 0.91 |
| TEX10 | 0.90 | 0.93 |
| CCT6A | 0.90 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.27 | -0.72 | -0.88 |
| AF347015.33 | -0.72 | -0.89 |
| MT-CO2 | -0.71 | -0.87 |
| HLA-F | -0.71 | -0.82 |
| AF347015.31 | -0.70 | -0.86 |
| MT-CYB | -0.70 | -0.87 |
| AF347015.8 | -0.69 | -0.88 |
| HEPN1 | -0.69 | -0.81 |
| AF347015.15 | -0.68 | -0.87 |
| AIFM3 | -0.68 | -0.78 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID PI3KCI PATHWAY | 49 | 40 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| WANG BARRETTS ESOPHAGUS DN | 25 | 13 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE LATE DN | 32 | 22 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| TAKAO RESPONSE TO UVB RADIATION UP | 86 | 55 | All SZGR 2.0 genes in this pathway |
| HU GENOTOXIN ACTION DIRECT VS INDIRECT 4HR | 37 | 22 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| LEE AGING MUSCLE UP | 45 | 33 | All SZGR 2.0 genes in this pathway |
| HILLION HMGA1B TARGETS | 92 | 68 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL OPTIMAL DEBULKING | 246 | 152 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| MUELLER COMMON TARGETS OF AML FUSIONS UP | 14 | 10 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |