Gene Page: ARRB1
Summary ?
| GeneID | 408 |
| Symbol | ARRB1 |
| Synonyms | ARB1|ARR1 |
| Description | arrestin, beta 1 |
| Reference | MIM:107940|HGNC:HGNC:711|Ensembl:ENSG00000137486|HPRD:00146|Vega:OTTHUMG00000165444 |
| Gene type | protein-coding |
| Map location | 11q13 |
| Pascal p-value | 0.045 |
| Fetal beta | -0.419 |
| DMG | 1 (# studies) |
| Support | CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0119 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02449461 | 11 | 75061838 | ARRB1 | 1.373E-4 | 0.271 | 0.031 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
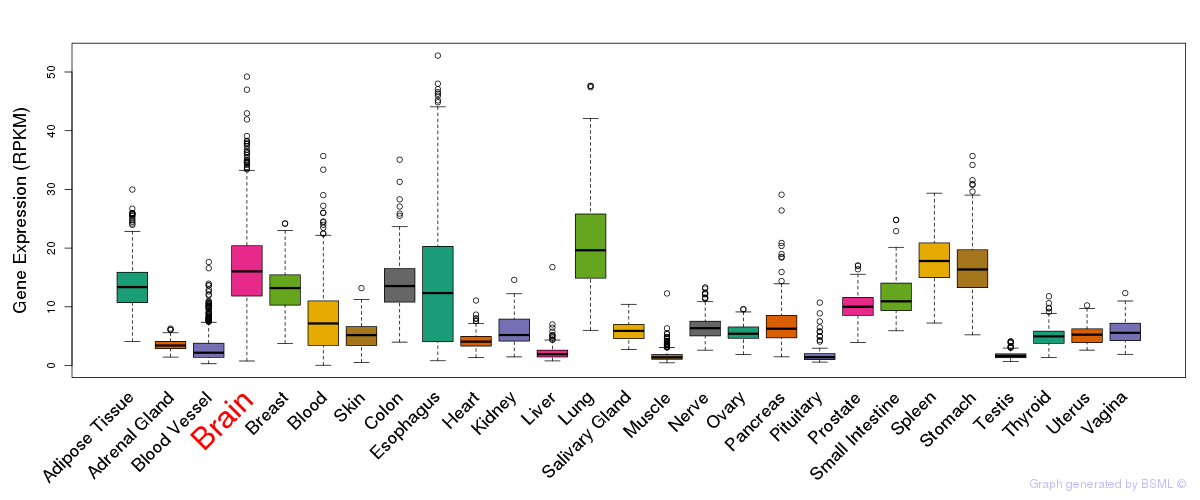
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SLC16A2 | 0.92 | 0.93 |
| PTPN9 | 0.91 | 0.94 |
| TGFBRAP1 | 0.91 | 0.93 |
| PUM1 | 0.90 | 0.92 |
| SARM1 | 0.90 | 0.92 |
| TRIO | 0.90 | 0.93 |
| TBC1D14 | 0.90 | 0.94 |
| ZCCHC14 | 0.90 | 0.94 |
| ZMYM3 | 0.90 | 0.92 |
| FOXJ2 | 0.90 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.76 | -0.84 |
| MT-CO2 | -0.75 | -0.85 |
| AF347015.27 | -0.72 | -0.81 |
| FXYD1 | -0.72 | -0.80 |
| AF347015.33 | -0.72 | -0.79 |
| C5orf53 | -0.71 | -0.74 |
| MT-CYB | -0.71 | -0.80 |
| AF347015.8 | -0.71 | -0.82 |
| S100B | -0.71 | -0.78 |
| HIGD1B | -0.70 | -0.82 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 17353931 |17620599 | |
| GO:0004857 | enzyme inhibitor activity | TAS | 2163110 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007165 | signal transduction | TAS | 9924018 | |
| GO:0007600 | sensory perception | IEA | - | |
| GO:0050896 | response to stimulus | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005834 | heterotrimeric G-protein complex | TAS | 10823817 | |
| GO:0005624 | membrane fraction | IDA | 10823817 | |
| GO:0005625 | soluble fraction | TAS | 9924018 | |
| GO:0005737 | cytoplasm | IDA | 10823817 | |
| GO:0005886 | plasma membrane | TAS | 9924018 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ADRB1 | ADRB1R | B1AR | BETA1AR | RHR | adrenergic, beta-1-, receptor | - | HPRD,BioGRID | 10862778 |
| ADRBK1 | BARK1 | BETA-ARK1 | FLJ16718 | GRK2 | adrenergic, beta, receptor kinase 1 | - | HPRD | 8885248 |
| AP2B1 | ADTB2 | AP105B | AP2-BETA | CLAPB1 | DKFZp781K0743 | adaptor-related protein complex 2, beta 1 subunit | - | HPRD | 11777907 |
| AP3B1 | ADTB3 | ADTB3A | HPS | HPS2 | PE | adaptor-related protein complex 3, beta 1 subunit | Affinity Capture-MS | BioGRID | 17353931 |
| ARF6 | DKFZp564M0264 | ADP-ribosylation factor 6 | - | HPRD,BioGRID | 11533043 |11867621 |
| ARR3 | ARRX | arrestin 3, retinal (X-arrestin) | Affinity Capture-MS | BioGRID | 17353931 |
| ARRB2 | ARB2 | ARR2 | BARR2 | DKFZp686L0365 | arrestin, beta 2 | Affinity Capture-MS | BioGRID | 17353931 |
| BOP1 | KIAA0124 | block of proliferation 1 | Affinity Capture-MS | BioGRID | 17353931 |
| CCL14 | CC-1 | CC-3 | CKb1 | HCC-1 | HCC-3 | MCIF | NCC-2 | NCC2 | SCYA14 | SCYL2 | SY14 | chemokine (C-C motif) ligand 14 | Affinity Capture-MS | BioGRID | 17353931 |
| CCR2 | CC-CKR-2 | CCR2A | CCR2B | CD192 | CKR2 | CKR2A | CKR2B | CMKBR2 | MCP-1-R | chemokine (C-C motif) receptor 2 | - | HPRD | 9501202 |
| CCR5 | CC-CKR-5 | CCCKR5 | CD195 | CKR-5 | CKR5 | CMKBR5 | chemokine (C-C motif) receptor 5 | - | HPRD,BioGRID | 12065593 |
| CHD1 | DKFZp686E2337 | chromodomain helicase DNA binding protein 1 | Affinity Capture-MS | BioGRID | 17353931 |
| CLTC | CHC | CHC17 | CLH-17 | CLTCL2 | Hc | KIAA0034 | clathrin, heavy chain (Hc) | - | HPRD | 9827808 |
| CMBL | FLJ23617 | carboxymethylenebutenolidase homolog (Pseudomonas) | Affinity Capture-MS | BioGRID | 17353931 |
| CSK | MGC117393 | c-src tyrosine kinase | - | HPRD,BioGRID | 10753943 |
| CSNK2A1 | CK2A1 | CKII | casein kinase 2, alpha 1 polypeptide | Affinity Capture-MS | BioGRID | 17353931 |
| CYTH2 | ARNO | CTS18 | CTS18.1 | PSCD2 | PSCD2L | SEC7L | Sec7p-L | Sec7p-like | cytohesin 2 | - | HPRD,BioGRID | 11533043 |
| DDX27 | DKFZp667N057 | FLJ12917 | FLJ20596 | FLJ22238 | HSPC259 | MGC1018 | MGC163147 | PP3241 | RHLP | Rrp3p | dJ686N3.1 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 27 | Affinity Capture-MS | BioGRID | 17353931 |
| DVL1 | DVL | MGC54245 | dishevelled, dsh homolog 1 (Drosophila) | - | HPRD,BioGRID | 11742073 |
| DVL2 | - | dishevelled, dsh homolog 2 (Drosophila) | - | HPRD,BioGRID | 11742073 |
| FGR | FLJ43153 | MGC75096 | SRC2 | c-fgr | c-src2 | p55c-fgr | p58c-fgr | Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog | - | HPRD | 10973280 |
| GNMT | - | glycine N-methyltransferase | Two-hybrid | BioGRID | 16189514 |
| GNRHR | GNRHR1 | GRHR | LHRHR | LRHR | gonadotropin-releasing hormone receptor | - | HPRD | 11278883 |
| HDGF2 | MGC2641 | hepatoma-derived growth factor-related protein 2 | Affinity Capture-MS | BioGRID | 17353931 |
| IL8RB | CD182 | CDw128b | CMKAR2 | CXCR2 | IL8R2 | IL8RA | interleukin 8 receptor, beta | - | HPRD,BioGRID | 11170396 |
| JAK1 | JAK1A | JAK1B | JTK3 | Janus kinase 1 (a protein tyrosine kinase) | Affinity Capture-MS | BioGRID | 17353931 |
| KHK | - | ketohexokinase (fructokinase) | Affinity Capture-MS | BioGRID | 17353931 |
| KPNA3 | IPOA4 | SRP1gamma | SRP4 | hSRP1 | karyopherin alpha 3 (importin alpha 4) | Affinity Capture-MS | BioGRID | 17353931 |
| MAPK10 | FLJ12099 | FLJ33785 | JNK3 | JNK3A | MGC50974 | PRKM10 | p493F12 | p54bSAPK | mitogen-activated protein kinase 10 | Affinity Capture-Western | BioGRID | 11356842 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | Affinity Capture-Western | BioGRID | 12538596 |
| NFKBIA | IKBA | MAD-3 | NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | Beta-Arr1 interacts with I-kappa-B-alpha to prevent the phosphorylation and degradation of I-kappa-B-alpha. | BIND | 15125834 |
| NOLC1 | KIAA0035 | NOPP130 | NOPP140 | NS5ATP13 | P130 | nucleolar and coiled-body phosphoprotein 1 | Affinity Capture-MS | BioGRID | 17353931 |
| NSF | SKD2 | N-ethylmaleimide-sensitive factor | - | HPRD,BioGRID | 10196135 |
| OPRD1 | OPRD | opioid receptor, delta 1 | - | HPRD,BioGRID | 11259507 |
| PDE4D | DPDE3 | HSPDE4D | PDE4DN2 | STRK1 | phosphodiesterase 4D, cAMP-specific (phosphodiesterase E3 dunce homolog, Drosophila) | The amino-terminus of PDE4D5 interacts with Beta-Arrestin 1. | BIND | 14500724 |
| PES1 | PES | pescadillo homolog 1, containing BRCT domain (zebrafish) | Affinity Capture-MS | BioGRID | 17353931 |
| PTH1R | MGC138426 | MGC138452 | PTHR | PTHR1 | parathyroid hormone 1 receptor | - | HPRD | 12220636 |
| PTHLH | HHM | MGC14611 | PLP | PTHR | PTHRP | parathyroid hormone-like hormone | - | HPRD,BioGRID | 12220636 |
| RALGDS | FLJ20922 | RGF | RalGEF | ral guanine nucleotide dissociation stimulator | - | HPRD,BioGRID | 12105416 |
| RPL7L1 | MGC62004 | dJ475N16.4 | ribosomal protein L7-like 1 | Affinity Capture-MS | BioGRID | 17353931 |
| RTF1 | CDG1N | GTL7 | KIAA0252 | Rtf1, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
| SLC9A5 | NHE5 | solute carrier family 9 (sodium/hydrogen exchanger), member 5 | An unspecified isoform of Beta-Arr1 interacts with NHE5. | BIND | 15699339 |
| TCOF1 | MFD1 | treacle | Treacher Collins-Franceschetti syndrome 1 | Affinity Capture-MS | BioGRID | 17353931 |
| TRHR | MGC141920 | thyrotropin-releasing hormone receptor | - | HPRD | 12393857 |
| ZBTB43 | ZBTB22B | ZNF-X | ZNF297B | zinc finger and BTB domain containing 43 | Two-hybrid | BioGRID | 16189514 |
| ZRANB2 | DKFZp686J1831 | DKFZp686N09117 | FLJ41119 | ZIS | ZIS1 | ZIS2 | ZNF265 | zinc finger, RAN-binding domain containing 2 | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG CHEMOKINE SIGNALING PATHWAY | 190 | 128 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AGPCR PATHWAY | 13 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BARR MAPK PATHWAY | 12 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BARRESTIN SRC PATHWAY | 15 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BARRESTIN PATHWAY | 10 | 8 | All SZGR 2.0 genes in this pathway |
| PID ARF6 PATHWAY | 35 | 27 | All SZGR 2.0 genes in this pathway |
| PID IL8 CXCR2 PATHWAY | 34 | 26 | All SZGR 2.0 genes in this pathway |
| PID CXCR3 PATHWAY | 43 | 34 | All SZGR 2.0 genes in this pathway |
| PID THROMBIN PAR1 PATHWAY | 43 | 32 | All SZGR 2.0 genes in this pathway |
| PID IL8 CXCR1 PATHWAY | 28 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME MEMBRANE TRAFFICKING | 129 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANS GOLGI NETWORK VESICLE BUDDING | 60 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | 27 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH1 | 70 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME LYSOSOME VESICLE BIOGENESIS | 23 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | 53 | 27 | All SZGR 2.0 genes in this pathway |
| REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | 32 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH | 103 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| DAVICIONI PAX FOXO1 SIGNATURE IN ARMS UP | 59 | 38 | All SZGR 2.0 genes in this pathway |
| IGARASHI ATF4 TARGETS DN | 90 | 65 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| DAVICIONI RHABDOMYOSARCOMA PAX FOXO1 FUSION UP | 64 | 37 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN UP | 184 | 125 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS UP | 306 | 188 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN | 384 | 230 | All SZGR 2.0 genes in this pathway |
| ODONNELL TFRC TARGETS UP | 456 | 228 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL DN | 186 | 107 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 AND TP63 TARGETS | 205 | 145 | All SZGR 2.0 genes in this pathway |
| EBAUER TARGETS OF PAX3 FOXO1 FUSION DN | 48 | 31 | All SZGR 2.0 genes in this pathway |
| SCHEIDEREIT IKK INTERACTING PROTEINS | 58 | 45 | All SZGR 2.0 genes in this pathway |
| YORDY RECIPROCAL REGULATION BY ETS1 AND SP100 UP | 25 | 11 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| GROSS ELK3 TARGETS UP | 27 | 16 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 UP | 209 | 139 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 ONLY DN | 44 | 30 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 120 HELA | 69 | 47 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 11Q12 Q14 AMPLICON | 158 | 93 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY MUTATED AND AMPLIFIED IN BREAST CANCER | 94 | 60 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP | 282 | 183 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS UP | 153 | 107 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| HOFFMANN IMMATURE TO MATURE B LYMPHOCYTE UP | 43 | 34 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A4 | 196 | 124 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| NIELSEN GIST | 98 | 66 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA NEURAL | 129 | 85 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN | 374 | 217 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| LIU IL13 MEMORY MODEL UP | 17 | 12 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38 | 337 | 236 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA S UP | 297 | 194 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-22 | 429 | 436 | 1A,m8 | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-379 | 107 | 113 | 1A | hsa-miR-379brain | UGGUAGACUAUGGAACGUA |
| miR-431 | 380 | 386 | 1A | hsa-miR-431 | UGUCUUGCAGGCCGUCAUGCA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.