Gene Page: PPP1R12B
Summary ?
| GeneID | 4660 |
| Symbol | PPP1R12B |
| Synonyms | MYPT2|PP1bp55 |
| Description | protein phosphatase 1 regulatory subunit 12B |
| Reference | MIM:603768|HGNC:HGNC:7619|Ensembl:ENSG00000077157|HPRD:04794|Vega:OTTHUMG00000041393 |
| Gene type | protein-coding |
| Map location | 1q32.1 |
| Fetal beta | 0.85 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08990198 | 1 | 202318195 | PPP1R12B | 6.81E-11 | -0.027 | 4.34E-7 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
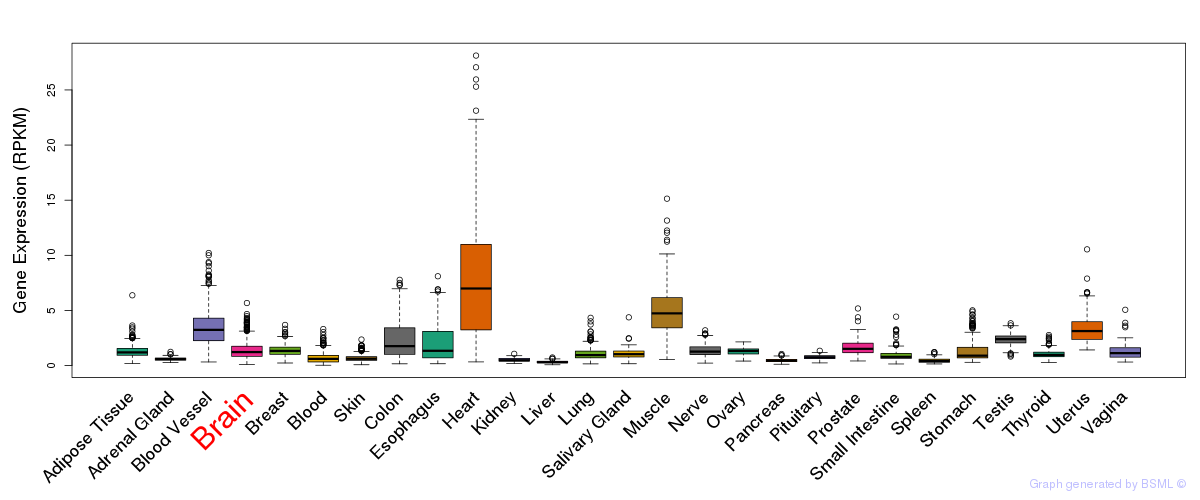
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ZMYM4 | 0.97 | 0.98 |
| WDR3 | 0.97 | 0.97 |
| KIAA1429 | 0.97 | 0.97 |
| SMEK2 | 0.97 | 0.96 |
| XRCC5 | 0.97 | 0.98 |
| SRP72 | 0.96 | 0.96 |
| CAND1 | 0.96 | 0.97 |
| CRNKL1 | 0.96 | 0.97 |
| MATR3 | 0.96 | 0.97 |
| WDR48 | 0.96 | 0.97 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.75 | -0.89 |
| MT-CO2 | -0.75 | -0.88 |
| AF347015.31 | -0.74 | -0.87 |
| AF347015.33 | -0.73 | -0.86 |
| IFI27 | -0.72 | -0.87 |
| AF347015.27 | -0.72 | -0.85 |
| AIFM3 | -0.72 | -0.79 |
| HLA-F | -0.72 | -0.78 |
| MT-CYB | -0.71 | -0.84 |
| AF347015.8 | -0.71 | -0.87 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG VASCULAR SMOOTH MUSCLE CONTRACTION | 115 | 81 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CCR3 PATHWAY | 23 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA INTEGRIN PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MYOSIN PATHWAY | 31 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RAC1 PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RHO PATHWAY | 32 | 23 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PAR1 PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL DN | 244 | 153 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| TOMLINS PROSTATE CANCER DN | 40 | 33 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| STOSSI RESPONSE TO ESTRADIOL | 50 | 35 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| KIM GASTRIC CANCER CHEMOSENSITIVITY | 103 | 64 | All SZGR 2.0 genes in this pathway |
| NIELSEN LEIOMYOSARCOMA CNN1 UP | 19 | 13 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL UP | 125 | 68 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS DN | 306 | 191 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP D | 280 | 158 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| DUAN PRDM5 TARGETS | 79 | 52 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |