Gene Page: NDUFA10
Summary ?
| GeneID | 4705 |
| Symbol | NDUFA10 |
| Synonyms | CI-42KD|CI-42k |
| Description | NADH:ubiquinone oxidoreductase subunit A10 |
| Reference | MIM:603835|HGNC:HGNC:7684|Ensembl:ENSG00000130414|HPRD:11950|Vega:OTTHUMG00000133350 |
| Gene type | protein-coding |
| Map location | 2q37.3 |
| Pascal p-value | 0.51 |
| Sherlock p-value | 0.004 |
| Fetal beta | -0.926 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum Cortex Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Meta |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_mitochondria G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03707784 | 2 | 240965007 | NDUFA10 | 1.42E-9 | -0.015 | 1.38E-6 | DMG:Jaffe_2016 |
| cg07584077 | 2 | 240965270 | NDUFA10 | 9.01E-9 | 0.008 | 4.11E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10166776 | 2 | 240967312 | NDUFA10 | ENSG00000130414.7 | 9.5827E-7 | 0.03 | -2493 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
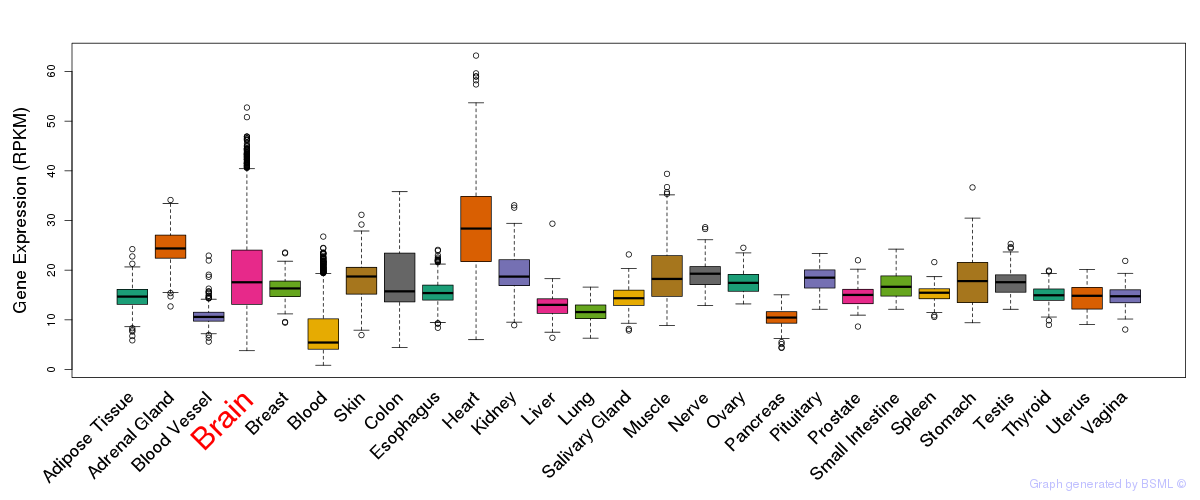
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TMEM141 | 0.87 | 0.69 |
| SNRNP25 | 0.86 | 0.78 |
| C7orf59 | 0.85 | 0.71 |
| C17orf90 | 0.85 | 0.69 |
| MRPL2 | 0.85 | 0.80 |
| ZNF593 | 0.84 | 0.69 |
| PIN1 | 0.84 | 0.80 |
| NDUFA2 | 0.84 | 0.67 |
| MRPS11 | 0.83 | 0.68 |
| SRA1 | 0.83 | 0.66 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC005035.1 | -0.36 | -0.38 |
| CDC42EP4 | -0.35 | -0.42 |
| MAP4K4 | -0.35 | -0.36 |
| AC005332.1 | -0.35 | -0.29 |
| ZNF652 | -0.33 | -0.29 |
| THOC2 | -0.33 | -0.32 |
| SBF2 | -0.33 | -0.27 |
| CDH5 | -0.33 | -0.32 |
| NEO1 | -0.32 | -0.29 |
| USP24 | -0.32 | -0.28 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG OXIDATIVE PHOSPHORYLATION | 135 | 73 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| KEGG PARKINSONS DISEASE | 133 | 78 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| REACTOME TCA CYCLE AND RESPIRATORY ELECTRON TRANSPORT | 141 | 85 | All SZGR 2.0 genes in this pathway |
| REACTOME RESPIRATORY ELECTRON TRANSPORT | 79 | 44 | All SZGR 2.0 genes in this pathway |
| REACTOME RESPIRATORY ELECTRON TRANSPORT ATP SYNTHESIS BY CHEMIOSMOTIC COUPLING AND HEAT PRODUCTION BY UNCOUPLING PROTEINS | 98 | 56 | All SZGR 2.0 genes in this pathway |
| PICCALUGA ANGIOIMMUNOBLASTIC LYMPHOMA DN | 136 | 86 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| MOOTHA VOXPHOS | 87 | 51 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| CHANG IMMORTALIZED BY HPV31 UP | 84 | 55 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| WONG MITOCHONDRIA GENE MODULE | 217 | 122 | All SZGR 2.0 genes in this pathway |
| MATTHEWS SKIN CARCINOGENESIS VIA JUN | 17 | 10 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |