Gene Page: NMBR
Summary ?
| GeneID | 4829 |
| Symbol | NMBR |
| Synonyms | BB1 |
| Description | neuromedin B receptor |
| Reference | MIM:162341|HGNC:HGNC:7843|Ensembl:ENSG00000135577|HPRD:01211|Vega:OTTHUMG00000015704 |
| Gene type | protein-coding |
| Map location | 6q24.1 |
| Pascal p-value | 0.142 |
| Fetal beta | 0.227 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Expression | Meta-analysis of gene expression | P value: 2.951 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
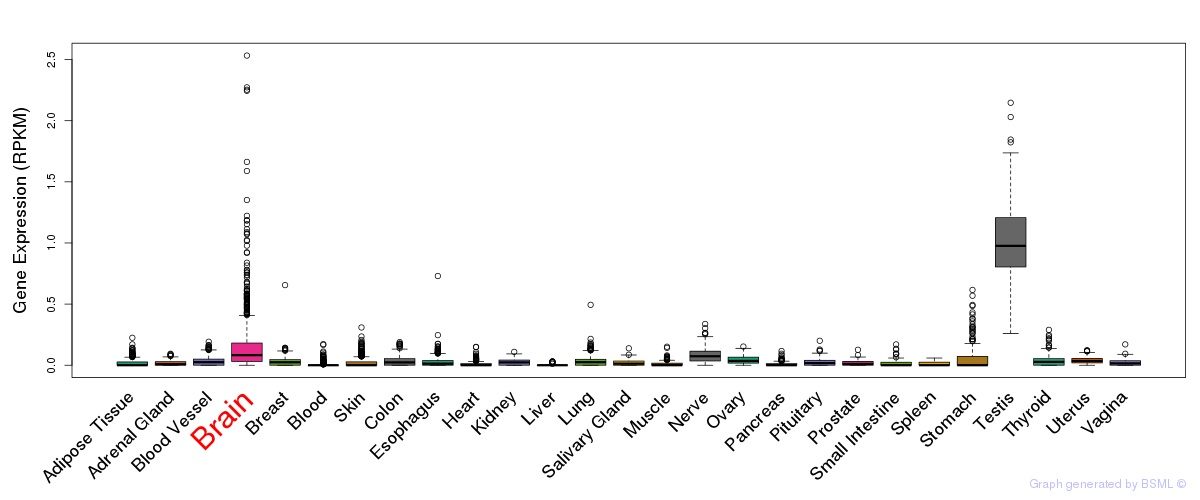
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DECR1 | 0.69 | 0.64 |
| ETFA | 0.69 | 0.59 |
| USP18 | 0.69 | 0.61 |
| AKR7A2 | 0.69 | 0.65 |
| TTC38 | 0.68 | 0.53 |
| LGTN | 0.68 | 0.62 |
| C2orf28 | 0.68 | 0.57 |
| ACAA2 | 0.67 | 0.66 |
| TMEM149 | 0.67 | 0.59 |
| DHRS3 | 0.67 | 0.66 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MEF2C | -0.43 | -0.52 |
| ARHGAP20 | -0.42 | -0.48 |
| DOCK9 | -0.41 | -0.46 |
| FBXW7 | -0.41 | -0.48 |
| HIVEP2 | -0.40 | -0.45 |
| CUX2 | -0.40 | -0.46 |
| C1orf115 | -0.39 | -0.47 |
| SLITRK5 | -0.39 | -0.46 |
| LRFN2 | -0.39 | -0.44 |
| AC016910.1 | -0.39 | -0.46 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| GO:0004946 | bombesin receptor activity | TAS | 7838118 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007200 | G-protein signaling, coupled to IP3 second messenger (phospholipase C activating) | TAS | 7838118 | |
| GO:0007165 | signal transduction | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005886 | plasma membrane | TAS | 7838118 | |
| GO:0005887 | integral to plasma membrane | TAS | 8391296 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| REACTOME GASTRIN CREB SIGNALLING PATHWAY VIA PKC AND MAPK | 205 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME PEPTIDE LIGAND BINDING RECEPTORS | 188 | 108 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS A1 RHODOPSIN LIKE RECEPTORS | 305 | 177 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA Q SIGNALLING EVENTS | 184 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| MCCLUNG DELTA FOSB TARGETS 2WK | 48 | 36 | All SZGR 2.0 genes in this pathway |