Gene Page: NODAL
Summary ?
| GeneID | 4838 |
| Symbol | NODAL |
| Synonyms | HTX5 |
| Description | nodal growth differentiation factor |
| Reference | MIM:601265|HGNC:HGNC:7865|HPRD:08370| |
| Gene type | protein-coding |
| Map location | 10q22.1 |
| Pascal p-value | 0.101 |
| Fetal beta | 0.193 |
| eGene | Caudate basal ganglia Nucleus accumbens basal ganglia Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizotypy,schizophrenias,schizotypal | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
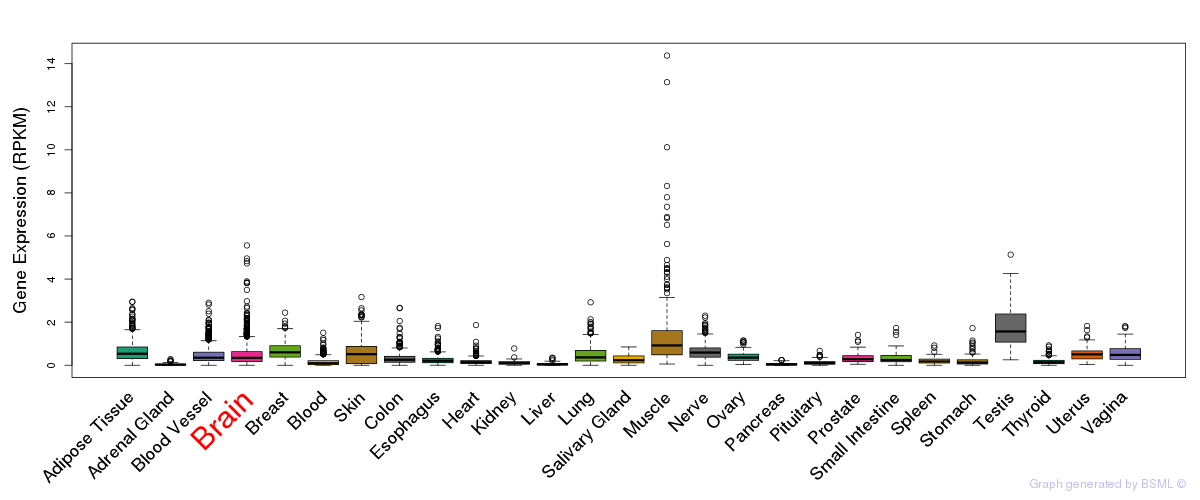
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HNRNPC | 0.95 | 0.93 |
| NAP1L1 | 0.94 | 0.93 |
| CPSF3 | 0.94 | 0.92 |
| TINP1 | 0.94 | 0.91 |
| MKI67IP | 0.93 | 0.94 |
| DCAF13 | 0.93 | 0.87 |
| HAUS1 | 0.93 | 0.81 |
| CCT2 | 0.92 | 0.91 |
| RBMX | 0.92 | 0.92 |
| EIF3E | 0.92 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.60 | -0.74 |
| AC018755.7 | -0.60 | -0.72 |
| SLC9A3R2 | -0.59 | -0.60 |
| AF347015.33 | -0.59 | -0.79 |
| MT-CO2 | -0.59 | -0.77 |
| MT-CYB | -0.58 | -0.77 |
| AF347015.31 | -0.57 | -0.75 |
| TINAGL1 | -0.57 | -0.74 |
| AF347015.27 | -0.57 | -0.75 |
| IFI27 | -0.57 | -0.77 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG TGF BETA SIGNALING PATHWAY | 86 | 64 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NODAL | 18 | 11 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| TESAR ALK TARGETS HUMAN ES 5D DN | 7 | 5 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| KOHOUTEK CCNT2 TARGETS | 58 | 35 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| WARTERS RESPONSE TO IR SKIN | 83 | 44 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |