Gene Page: NOS1
Summary ?
| GeneID | 4842 |
| Symbol | NOS1 |
| Synonyms | IHPS1|N-NOS|NC-NOS|NOS|bNOS|nNOS |
| Description | nitric oxide synthase 1 |
| Reference | MIM:163731|HGNC:HGNC:7872|Ensembl:ENSG00000089250|HPRD:01226|Vega:OTTHUMG00000137376 |
| Gene type | protein-coding |
| Map location | 12q24.22 |
| Pascal p-value | 5.53E-6 |
| Fetal beta | -0.854 |
| DMG | 1 (# studies) |
| eGene | Cortex Hippocampus |
| Support | G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_PocklingtonH1 G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 2 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg21273407 | 12 | 117799447 | NOS1 | 3.537E-4 | -0.428 | 0.042 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
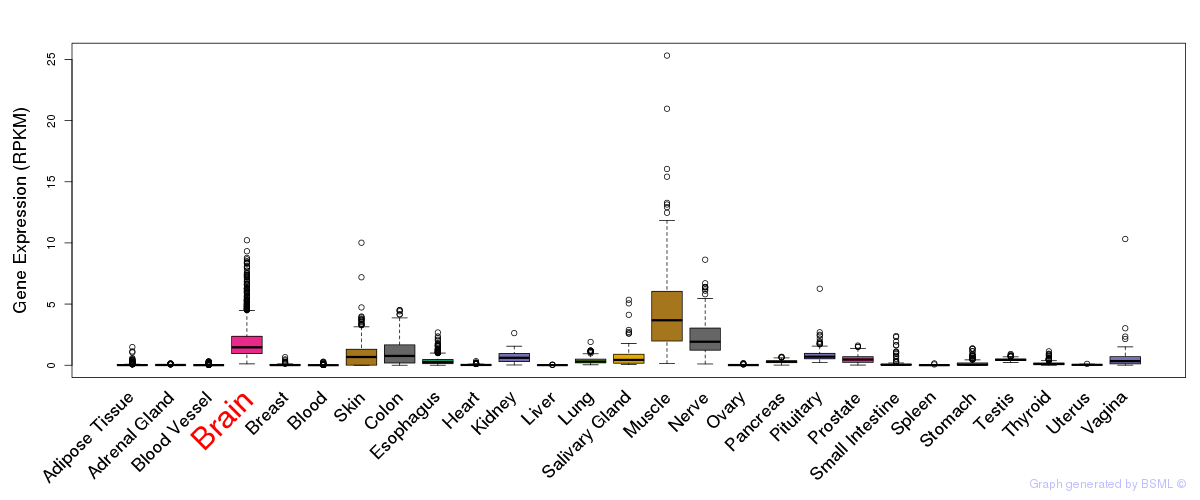
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005506 | iron ion binding | IEA | - | |
| GO:0005516 | calmodulin binding | IEA | - | |
| GO:0004517 | nitric-oxide synthase activity | IEA | - | |
| GO:0020037 | heme binding | IEA | - | |
| GO:0010181 | FMN binding | IEA | - | |
| GO:0009055 | electron carrier activity | IEA | - | |
| GO:0016491 | oxidoreductase activity | IEA | - | |
| GO:0034617 | tetrahydrobiopterin binding | NAS | 7488039 | |
| GO:0034618 | arginine binding | TAS | 17029414 | |
| GO:0046870 | cadmium ion binding | ISS | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| GO:0050660 | FAD binding | IEA | - | |
| GO:0050661 | NADP binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0042136 | neurotransmitter biosynthetic process | TAS | neuron, axon, Synap, Neurotransmitter (GO term level: 9) | 7545544 |
| GO:0001666 | response to hypoxia | IEP | 16276418 | |
| GO:0006527 | arginine catabolic process | IC | 7545544 | |
| GO:0010523 | negative regulation of calcium ion transport into cytosol | TAS | 17568574 | |
| GO:0009408 | response to heat | IDA | 18048451 | |
| GO:0007520 | myoblast fusion | TAS | 7545544 | |
| GO:0006809 | nitric oxide biosynthetic process | IEA | - | |
| GO:0033555 | multicellular organismal response to stress | IMP | 18391107 | |
| GO:0045909 | positive regulation of vasodilation | IDA | 18048451 | |
| GO:0045909 | positive regulation of vasodilation | IMP | 18391107 | |
| GO:0055114 | oxidation reduction | IEA | - | |
| GO:0055117 | regulation of cardiac muscle contraction | TAS | 9892689 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0042995 | cell projection | IEA | axon (GO term level: 4) | - |
| GO:0001917 | photoreceptor inner segment | ISS | 17027776 | |
| GO:0005856 | cytoskeleton | ISS | 7545544 | |
| GO:0005737 | cytoplasm | TAS | 17892502 | |
| GO:0048471 | perinuclear region of cytoplasm | ISS | 17027776 | |
| GO:0042383 | sarcolemma | IDA | 7545544 | |
| GO:0016529 | sarcoplasmic reticulum | IDA | 9892689 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARG1 | - | arginase, liver | - | HPRD | 11849441 |
| BDKRB2 | B2R | BK-2 | BK2 | BKR2 | BRB2 | DKFZp686O088 | bradykinin receptor B2 | - | HPRD,BioGRID | 10681501 |
| CAV3 | LGMD1C | LQT9 | MGC126100 | MGC126101 | MGC126129 | VIP-21 | VIP21 | caveolin 3 | - | HPRD | 9353265 |
| DLG2 | DKFZp781D1854 | DKFZp781E0954 | FLJ37266 | MGC131811 | PSD-93 | discs, large homolog 2, chapsyn-110 (Drosophila) | - | HPRD,BioGRID | 8625413 |
| DLG4 | FLJ97752 | FLJ98574 | PSD95 | SAP90 | discs, large homolog 4 (Drosophila) | - | HPRD,BioGRID | 8625413 |
| DYNLL1 | DLC1 | DLC8 | DNCL1 | DNCLC1 | LC8 | LC8a | MGC126137 | MGC126138 | PIN | hdlc1 | dynein, light chain, LC8-type 1 | - | HPRD | 8864115 |
| HMOX1 | HO-1 | HSP32 | bK286B10 | heme oxygenase (decycling) 1 | - | HPRD,BioGRID | 11849436 |
| NOS1AP | 6330408P19Rik | CAPON | MGC138500 | nitric oxide synthase 1 (neuronal) adaptor protein | - | HPRD,BioGRID | 9459447 |
| NOSIP | CGI-25 | nitric oxide synthase interacting protein | - | HPRD | 15548660 |
| PTPN6 | HCP | HCPH | HPTP1C | PTP-1C | SH-PTP1 | SHP-1 | SHP-1L | SHP1 | protein tyrosine phosphatase, non-receptor type 6 | - | HPRD,BioGRID | 11511520 |
| PTPRN | FLJ16131 | IA-2 | IA-2/PTP | IA2 | ICA512 | R-PTP-N | protein tyrosine phosphatase, receptor type, N | - | HPRD | 11043403 |
| PUM2 | FLJ36528 | KIAA0235 | MGC138251 | MGC138253 | PUMH2 | PUML2 | pumilio homolog 2 (Drosophila) | Affinity Capture-Western | BioGRID | 12690449 |
| RASD1 | AGS1 | DEXRAS1 | MGC:26290 | RAS, dexamethasone-induced 1 | Reconstituted Complex | BioGRID | 11086993 |
| SNTA1 | SNT1 | TACIP1 | dJ1187J4.5 | syntrophin, alpha 1 (dystrophin-associated protein A1, 59kDa, acidic component) | - | HPRD | 11747091 |
| SNTA1 | SNT1 | TACIP1 | dJ1187J4.5 | syntrophin, alpha 1 (dystrophin-associated protein A1, 59kDa, acidic component) | Reconstituted Complex | BioGRID | 9412493 |
| SYN1 | SYN1a | SYN1b | SYNI | synapsin I | Reconstituted Complex | BioGRID | 11867766 |
| ZDHHC23 | MGC42530 | NIDD | zinc finger, DHHC-type containing 23 | Affinity Capture-Western Reconstituted Complex | BioGRID | 15105416 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ARGININE AND PROLINE METABOLISM | 54 | 39 | All SZGR 2.0 genes in this pathway |
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM DEPRESSION | 70 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| KEGG AMYOTROPHIC LATERAL SCLEROSIS ALS | 53 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NOS1 PATHWAY | 24 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | 33 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME NITRIC OXIDE STIMULATES GUANYLATE CYCLASE | 25 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET HOMEOSTASIS | 78 | 49 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER C | 113 | 47 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT UP | 165 | 100 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| XU GH1 EXOGENOUS TARGETS UP | 85 | 50 | All SZGR 2.0 genes in this pathway |
| LIU SMARCA4 TARGETS | 64 | 39 | All SZGR 2.0 genes in this pathway |
| XU GH1 AUTOCRINE TARGETS UP | 268 | 157 | All SZGR 2.0 genes in this pathway |
| RIGGI EWING SARCOMA PROGENITOR UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| MATZUK EARLY ANTRAL FOLLICLE | 13 | 11 | All SZGR 2.0 genes in this pathway |
| CHO NR4A1 TARGETS | 33 | 22 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS HCP WITH H3 UNMETHYLATED | 80 | 50 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES HCP WITH H3 UNMETHYLATED | 63 | 31 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3 UNMETHYLATED | 228 | 119 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE M G1 | 148 | 95 | All SZGR 2.0 genes in this pathway |