Gene Page: PAFAH1B1
Summary ?
| GeneID | 5048 |
| Symbol | PAFAH1B1 |
| Synonyms | LIS1|LIS2|MDCR|MDS|PAFAH |
| Description | platelet activating factor acetylhydrolase 1b regulatory subunit 1 |
| Reference | MIM:601545|HGNC:HGNC:8574|Ensembl:ENSG00000007168|HPRD:03329|Vega:OTTHUMG00000177574 |
| Gene type | protein-coding |
| Map location | 17p13.3 |
| Pascal p-value | 0.043 |
| Sherlock p-value | 0.446 |
| Fetal beta | 0.455 |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 8 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.1894 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
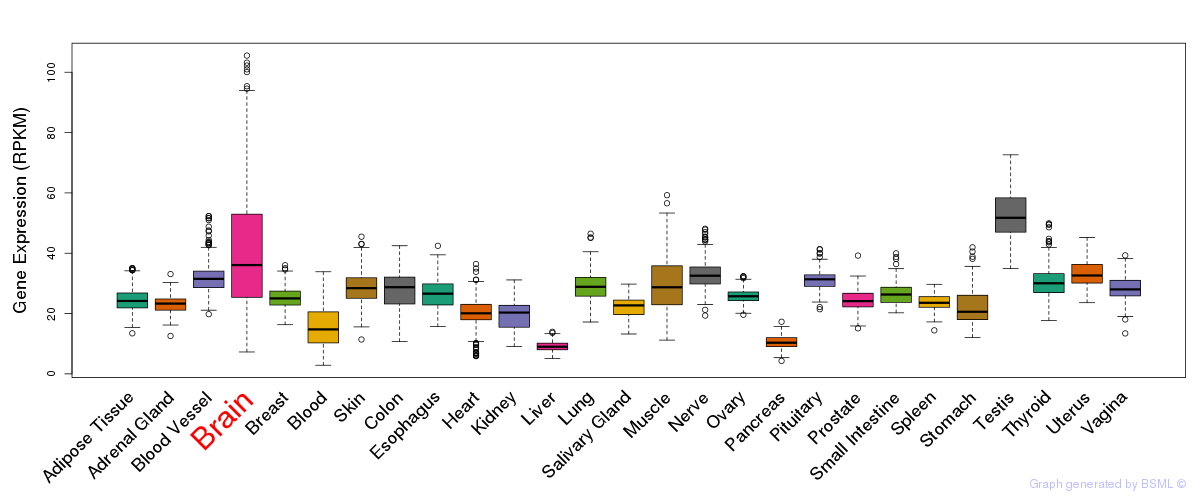
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CDCA7L | 0.87 | 0.61 |
| CDK2 | 0.86 | 0.48 |
| GLI3 | 0.86 | 0.68 |
| WEE1 | 0.85 | 0.35 |
| HELLS | 0.85 | 0.25 |
| MELK | 0.85 | 0.27 |
| FANCI | 0.85 | 0.26 |
| TEAD2 | 0.85 | 0.40 |
| KIF23 | 0.85 | 0.19 |
| CCNB2 | 0.85 | 0.28 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ADAP1 | -0.34 | -0.28 |
| FXYD7 | -0.33 | -0.25 |
| P2RX5 | -0.32 | -0.31 |
| KCNF1 | -0.32 | -0.36 |
| PINK1 | -0.32 | -0.33 |
| KAZALD1 | -0.32 | -0.24 |
| RLTPR | -0.32 | -0.26 |
| ASPHD1 | -0.31 | -0.20 |
| CKMT1A | -0.31 | -0.29 |
| SULT4A1 | -0.31 | -0.40 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008201 | heparin binding | ISS | 8028668 | |
| GO:0008017 | microtubule binding | IEA | - | |
| GO:0051219 | phosphoprotein binding | ISS | - | |
| GO:0042803 | protein homodimerization activity | IEA | - | |
| GO:0043274 | phospholipase binding | ISS | 8028668 | |
| GO:0034452 | dynactin binding | ISS | 11056532 | |
| GO:0045502 | dynein binding | IDA | 11889140 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007405 | neuroblast proliferation | IEA | neuron (GO term level: 8) | - |
| GO:0001764 | neuron migration | IEA | neuron (GO term level: 8) | - |
| GO:0007268 | synaptic transmission | IEA | neuron, Synap, Neurotransmitter (GO term level: 6) | - |
| GO:0021540 | corpus callosum morphogenesis | IMP | neuron, axon (GO term level: 15) | 9063735 |
| GO:0048854 | brain morphogenesis | IMP | Brain (GO term level: 8) | 9063735 |
| GO:0008090 | retrograde axon cargo transport | IEA | axon (GO term level: 9) | - |
| GO:0021819 | layer formation in the cerebral cortex | ISS | Glial (GO term level: 15) | - |
| GO:0007399 | nervous system development | IEA | neurite (GO term level: 5) | - |
| GO:0000132 | establishment of mitotic spindle orientation | IMP | 11056532 | |
| GO:0000226 | microtubule cytoskeleton organization | IEA | - | |
| GO:0001675 | acrosome formation | IEA | - | |
| GO:0016042 | lipid catabolic process | IEA | - | |
| GO:0007049 | cell cycle | IEA | - | |
| GO:0007067 | mitosis | IEA | - | |
| GO:0008344 | adult locomotory behavior | IEA | - | |
| GO:0007611 | learning or memory | IEA | - | |
| GO:0006810 | transport | IEA | - | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0030036 | actin cytoskeleton organization | ISS | 10729324 | |
| GO:0021987 | cerebral cortex development | IEA | - | |
| GO:0050885 | neuromuscular process controlling balance | IEA | - | |
| GO:0021766 | hippocampus development | IEA | - | |
| GO:0032319 | regulation of Rho GTPase activity | ISS | 10729324 | |
| GO:0051301 | cell division | IEA | - | |
| GO:0047496 | vesicle transport along microtubule | IEA | - | |
| GO:0046469 | platelet activating factor metabolic process | ISS | 8028668 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000235 | astral microtubule | IDA | 11940666 | |
| GO:0000776 | kinetochore | IDA | 11056532 |11889140 |11940666 | |
| GO:0005813 | centrosome | IEA | - | |
| GO:0005819 | spindle | IEA | - | |
| GO:0005829 | cytosol | ISS | 8028668 | |
| GO:0005874 | microtubule | IEA | - | |
| GO:0005875 | microtubule associated complex | IDA | 11889140 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0005938 | cell cortex | IDA | 11940666 | |
| GO:0048471 | perinuclear region of cytoplasm | ISS | - | |
| GO:0031513 | nonmotile primary cilium | ISS | - | |
| GO:0031512 | motile primary cilium | IEA | - | |
| GO:0031252 | cell leading edge | IEA | - | |
| GO:0031965 | nuclear membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CLIP1 | CLIP | CLIP-170 | CLIP170 | CYLN1 | MGC131604 | RSN | CAP-GLY domain containing linker protein 1 | - | HPRD,BioGRID | 11940666 |
| CSNK2A1 | CK2A1 | CKII | casein kinase 2, alpha 1 polypeptide | - | HPRD,BioGRID | 10491172 |
| DAB1 | - | disabled homolog 1 (Drosophila) | - | HPRD | 14578885 |
| DCTN1 | DAP-150 | DP-150 | HMN7B | P135 | dynactin 1 (p150, glued homolog, Drosophila) | - | HPRD,BioGRID | 11889140|14584027 |
| DCTN2 | DCTN50 | DYNAMITIN | RBP50 | dynactin 2 (p50) | Two-hybrid | BioGRID | 11889140 |
| DCX | DBCN | DC | LISX | SCLH | XLIS | doublecortin | - | HPRD,BioGRID | 11001923 |
| DNAI1 | CILD1 | ICS | MGC26204 | PCD | dynein, axonemal, intermediate chain 1 | - | HPRD | 11889140 |
| DYNC1H1 | DHC1 | DHC1a | DKFZp686P2245 | DNCH1 | DNCL | DNECL | DYHC | Dnchc1 | HL-3 | KIAA0325 | p22 | dynein, cytoplasmic 1, heavy chain 1 | Affinity Capture-Western Two-hybrid | BioGRID | 11889140 |
| HSPH1 | DKFZp686M05240 | HSP105 | HSP105A | HSP105B | KIAA0201 | NY-CO-25 | heat shock 105kDa/110kDa protein 1 | - | HPRD | 14733918 |
| KARS | KARS2 | KIAA0070 | KRS | lysyl-tRNA synthetase | Two-hybrid | BioGRID | 16169070 |
| NDE1 | FLJ20101 | HOM-TES-87 | NUDE | NUDE1 | nudE nuclear distribution gene E homolog 1 (A. nidulans) | - | HPRD | 11163258 |
| NDEL1 | DKFZp451M0318 | EOPA | MITAP1 | NUDEL | nudE nuclear distribution gene E homolog (A. nidulans)-like 1 | - | HPRD,BioGRID | 11163260 |
| NUDC | HNUDC | MNUDC | NPD011 | nuclear distribution gene C homolog (A. nidulans) | - | HPRD,BioGRID | 9601647 |11734602 |
| PAFAH1B1 | LIS1 | LIS2 | MDCR | MDS | PAFAH | platelet-activating factor acetylhydrolase, isoform Ib, alpha subunit 45kDa | - | HPRD,BioGRID | 11889140 |
| PAFAH1B2 | - | platelet-activating factor acetylhydrolase, isoform Ib, beta subunit 30kDa | - | HPRD,BioGRID | 10727864 |
| PAFAH1B3 | - | platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa | - | HPRD,BioGRID | 10727864 |
| TUBA1A | B-ALPHA-1 | FLJ25113 | LIS3 | TUBA3 | tubulin, alpha 1a | Reconstituted Complex | BioGRID | 9384577 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ETHER LIPID METABOLISM | 33 | 15 | All SZGR 2.0 genes in this pathway |
| PID REELIN PATHWAY | 29 | 29 | All SZGR 2.0 genes in this pathway |
| PID LIS1 PATHWAY | 28 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME RECRUITMENT OF MITOTIC CENTROSOME PROTEINS AND COMPLEXES | 66 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME LOSS OF NLP FROM MITOTIC CENTROSOMES | 59 | 38 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G2 G2 M PHASES | 81 | 50 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC PROMETAPHASE | 87 | 51 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER UP | 181 | 108 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS DN | 124 | 79 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BUYTAERT PHOTODYNAMIC THERAPY STRESS UP | 811 | 508 | All SZGR 2.0 genes in this pathway |
| SCHWAB TARGETS OF BMYB POLYMORPHIC VARIANTS DN | 17 | 12 | All SZGR 2.0 genes in this pathway |
| GOTZMANN EPITHELIAL TO MESENCHYMAL TRANSITION UP | 69 | 55 | All SZGR 2.0 genes in this pathway |
| GEORGES CELL CYCLE MIR192 TARGETS | 62 | 46 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 DN | 169 | 102 | All SZGR 2.0 genes in this pathway |
| GOLDRATH HOMEOSTATIC PROLIFERATION | 171 | 102 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| KAAB FAILED HEART ATRIUM DN | 141 | 99 | All SZGR 2.0 genes in this pathway |
| JIANG AGING CEREBRAL CORTEX UP | 36 | 27 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR DN | 91 | 64 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D6 | 37 | 25 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN | 128 | 93 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT DN | 165 | 106 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| LY AGING OLD DN | 56 | 35 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G6 | 153 | 112 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C5 | 46 | 36 | All SZGR 2.0 genes in this pathway |
| GENTILE UV LOW DOSE DN | 67 | 46 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 12HR DN | 101 | 64 | All SZGR 2.0 genes in this pathway |
| JIANG AGING HYPOTHALAMUS UP | 47 | 31 | All SZGR 2.0 genes in this pathway |
| OUILLETTE CLL 13Q14 DELETION UP | 74 | 40 | All SZGR 2.0 genes in this pathway |
| DE YY1 TARGETS DN | 92 | 64 | All SZGR 2.0 genes in this pathway |
| MATZUK SPERMATID DIFFERENTIATION | 37 | 26 | All SZGR 2.0 genes in this pathway |
| BONCI TARGETS OF MIR15A AND MIR16 1 | 91 | 75 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| RAMPON ENRICHED LEARNING ENVIRONMENT EARLY DN | 10 | 10 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 2739 | 2746 | 1A,m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-10 | 1040 | 1046 | m8 | hsa-miR-10a | UACCCUGUAGAUCCGAAUUUGUG |
| hsa-miR-10b | UACCCUGUAGAACCGAAUUUGU | ||||
| miR-101 | 181 | 187 | 1A | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-124/506 | 2549 | 2555 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-125/351 | 2362 | 2369 | 1A,m8 | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-130/301 | 2868 | 2874 | m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-144 | 180 | 187 | 1A,m8 | hsa-miR-144 | UACAGUAUAGAUGAUGUACUAG |
| miR-15/16/195/424/497 | 2005 | 2012 | 1A,m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG | ||||
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG | ||||
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-150 | 2287 | 2294 | 1A,m8 | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
| miR-151 | 202 | 208 | 1A | hsa-miR-151brain | ACUAGACUGAAGCUCCUUGAGG |
| miR-17-5p/20/93.mr/106/519.d | 2870 | 2877 | 1A,m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-181 | 3673 | 3679 | 1A | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-182 | 566 | 573 | 1A,m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-194 | 139 | 145 | 1A | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| miR-205 | 3555 | 3561 | m8 | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| miR-214 | 2304 | 2310 | m8 | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| miR-22 | 667 | 673 | 1A | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-223 | 3549 | 3555 | 1A | hsa-miR-223 | UGUCAGUUUGUCAAAUACCCC |
| miR-25/32/92/363/367 | 893 | 899 | m8 | hsa-miR-25brain | CAUUGCACUUGUCUCGGUCUGA |
| hsa-miR-32 | UAUUGCACAUUACUAAGUUGC | ||||
| hsa-miR-92 | UAUUGCACUUGUCCCGGCCUG | ||||
| hsa-miR-367 | AAUUGCACUUUAGCAAUGGUGA | ||||
| hsa-miR-92bSZ | UAUUGCACUCGUCCCGGCCUC | ||||
| miR-324-5p | 38 | 44 | 1A | hsa-miR-324-5p | CGCAUCCCCUAGGGCAUUGGUGU |
| miR-330 | 2638 | 2644 | m8 | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-339 | 1039 | 1045 | m8 | hsa-miR-339 | UCCCUGUCCUCCAGGAGCUCA |
| miR-376c | 431 | 438 | 1A,m8 | hsa-miR-376c | AACAUAGAGGAAAUUCCACG |
| miR-381 | 176 | 182 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-496 | 101 | 108 | 1A,m8 | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-503 | 2006 | 2012 | 1A | hsa-miR-503 | UAGCAGCGGGAACAGUUCUGCAG |
| miR-543 | 137 | 143 | 1A | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
| miR-96 | 567 | 573 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.