Gene Page: PAX6
Summary ?
| GeneID | 5080 |
| Symbol | PAX6 |
| Synonyms | AN|AN2|D11S812E|FVH1|MGDA|WAGR |
| Description | paired box 6 |
| Reference | MIM:607108|HGNC:HGNC:8620|Ensembl:ENSG00000007372|HPRD:06167| |
| Gene type | protein-coding |
| Map location | 11p13 |
| Pascal p-value | 0.048 |
| Sherlock p-value | 0.382 |
| Fetal beta | 1.225 |
| DMG | 2 (# studies) |
| eGene | Nucleus accumbens basal ganglia Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15778437 | 11 | 31839521 | PAX6 | 2.04E-5 | -0.414 | 0.016 | DMG:Wockner_2014 |
| cg17280740 | 11 | 31840862 | PAX6 | 5.882E-4 | 0.326 | 0.05 | DMG:Wockner_2014 |
| cg13023210 | 11 | 31821452 | PAX6 | 5.84E-9 | -0.026 | 3.16E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17392743 | chr1 | 54764240 | PAX6 | 5080 | 0.04 | trans | ||
| rs2502827 | chr1 | 176044216 | PAX6 | 5080 | 0.09 | trans | ||
| rs17572651 | chr1 | 218943612 | PAX6 | 5080 | 0 | trans | ||
| rs16829545 | chr2 | 151977407 | PAX6 | 5080 | 1.783E-10 | trans | ||
| rs3845734 | chr2 | 171125572 | PAX6 | 5080 | 0 | trans | ||
| rs1967327 | chr2 | 179314358 | PAX6 | 5080 | 0.19 | trans | ||
| rs7584986 | chr2 | 184111432 | PAX6 | 5080 | 5.069E-8 | trans | ||
| rs17762315 | chr5 | 76807576 | PAX6 | 5080 | 0.03 | trans | ||
| rs7729096 | chr5 | 76835927 | PAX6 | 5080 | 0.16 | trans | ||
| rs2393316 | chr10 | 59333070 | PAX6 | 5080 | 0.05 | trans | ||
| rs17104720 | chr14 | 77127308 | PAX6 | 5080 | 0.03 | trans | ||
| rs16955618 | chr15 | 29937543 | PAX6 | 5080 | 1.26E-19 | trans | ||
| rs1041786 | chr21 | 22617710 | PAX6 | 5080 | 0.04 | trans |
Section II. Transcriptome annotation
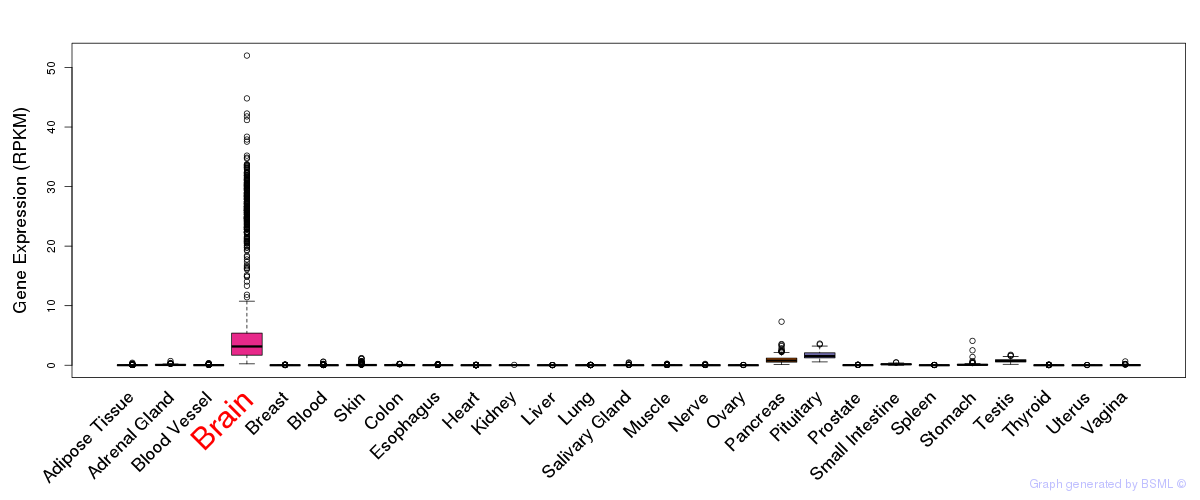
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003700 | transcription factor activity | TAS | 10441571 |10747901 | |
| GO:0005515 | protein binding | IPI | 16098226 | |
| GO:0043565 | sequence-specific DNA binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007417 | central nervous system development | TAS | Brain (GO term level: 6) | 10747901 |
| GO:0001654 | eye development | TAS | 10747901 | |
| GO:0006355 | regulation of transcription, DNA-dependent | IEA | - | |
| GO:0009887 | organ morphogenesis | TAS | 10441571 | |
| GO:0007601 | visual perception | TAS | 10441571 | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CDX2 | CDX-3 | CDX3 | caudal type homeobox 2 | - | HPRD,BioGRID | 10506141 |
| EN1 | - | engrailed homeobox 1 | Reconstituted Complex | BioGRID | 11069920 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | Affinity Capture-Western Reconstituted Complex | BioGRID | 10506141 |
| HOXB1 | HOX2 | HOX2I | Hox-2.9 | MGC116843 | MGC116844 | MGC116845 | homeobox B1 | - | HPRD,BioGRID | 11069920 |
| HOXB1 | HOX2 | HOX2I | Hox-2.9 | MGC116843 | MGC116844 | MGC116845 | homeobox B1 | Pax6 interacts with HoxB1. This interaction was modeled on a demonstrated interaction between zebrafish Pax6 and human HoxB1. | BIND | 11069920 |
| IPO13 | IMP13 | KAP13 | KIAA0724 | RANBP13 | importin 13 | Kap13 interacts with Pax6. | BIND | 15143176 |
| IPO13 | IMP13 | KAP13 | KIAA0724 | RANBP13 | importin 13 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 15143176 |16189514 |
| LHX2 | LH2 | MGC138390 | hLhx2 | LIM homeobox 2 | Reconstituted Complex | BioGRID | 11069920 |
| MAFG | MGC13090 | MGC20149 | v-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian) | - | HPRD,BioGRID | 11036080 |
| MITF | MI | WS2A | bHLHe32 | microphthalmia-associated transcription factor | - | HPRD,BioGRID | 11350962 |
| PAX6 | AN | AN2 | D11S812E | MGC17209 | MGDA | WAGR | paired box 6 | Affinity Capture-Western Reconstituted Complex | BioGRID | 11069920 |
| PBX1 | DKFZp686B09108 | MGC126627 | pre-B-cell leukemia homeobox 1 | Pax6 interacts with Pbx1. This interaction was modeled on a demonstrated interaction between zebrafish Pax6 and human Pbx1. | BIND | 11069920 |
| PKNOX1 | PREP1 | pkonx1c | PBX/knotted 1 homeobox 1 | Reconstituted Complex | BioGRID | 11069920 |
| PROX1 | - | prospero homeobox 1 | Reconstituted Complex | BioGRID | 11069920 |
| RAX | MCOP3 | RX | retina and anterior neural fold homeobox | - | HPRD,BioGRID | 11069920 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | Reconstituted Complex | BioGRID | 10359315 |
| SIX3 | HPE2 | SIX homeobox 3 | - | HPRD,BioGRID | 11069920 |
| SOX2 | ANOP3 | MCOPS3 | MGC2413 | SRY (sex determining region Y)-box 2 | - | HPRD,BioGRID | 11358870 |12710953 |12923055 |
| SOX3 | GHDX | MRGH | PHP | PHPX | SOXB | SRY (sex determining region Y)-box 3 | - | HPRD | 12710953 |
| TAF1 | BA2R | CCG1 | CCGS | DYT3 | KAT4 | N-TAF1 | NSCL2 | OF | P250 | TAF2A | TAFII250 | TAF1 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 250kDa | Affinity Capture-Western | BioGRID | 10359315 |
| TBP | GTF2D | GTF2D1 | MGC117320 | MGC126054 | MGC126055 | SCA17 | TFIID | TATA box binding protein | - | HPRD,BioGRID | 10359315 |
| VSX2 | CHX10 | HOX10 | MCOP2 | MCOPCB3 | RET1 | visual system homeobox 2 | Reconstituted Complex | BioGRID | 11069920 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MATURITY ONSET DIABETES OF THE YOUNG | 25 | 19 | All SZGR 2.0 genes in this pathway |
| PID CDC42 PATHWAY | 70 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF BETA CELL DEVELOPMENT | 30 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | 20 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRATION OF ENERGY METABOLISM | 120 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF INSULIN SECRETION | 93 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GIP | 14 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME INCRETIN SYNTHESIS SECRETION AND INACTIVATION | 22 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | 19 | 12 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER ESR1 DN | 48 | 28 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| YEMELYANOV GR TARGETS DN | 10 | 8 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| SCHLESINGER METHYLATED DE NOVO IN CANCER | 88 | 64 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES CORE NINE CORRELATED | 100 | 68 | All SZGR 2.0 genes in this pathway |
| TESAR ALK TARGETS HUMAN ES 5D UP | 5 | 5 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| WEBER METHYLATED IN COLON CANCER | 18 | 13 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 2 | 50 | 34 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED EPIGENETICALLY IN PANCREATIC CANCER | 49 | 30 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| TESAR ALK TARGETS EPISC 4D UP | 7 | 6 | All SZGR 2.0 genes in this pathway |
| TESAR ALK AND JAK TARGETS MOUSE ES D4 UP | 5 | 5 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| ZHOU PANCREATIC ENDOCRINE PROGENITOR | 14 | 11 | All SZGR 2.0 genes in this pathway |
| ZHOU PANCREATIC BETA CELL | 11 | 9 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| DUAN PRDM5 TARGETS | 79 | 52 | All SZGR 2.0 genes in this pathway |
| HUANG GATA2 TARGETS UP | 149 | 96 | All SZGR 2.0 genes in this pathway |
| WANG MLL TARGETS | 289 | 188 | All SZGR 2.0 genes in this pathway |
| ROLEF GLIS3 TARGETS | 37 | 29 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-129-5p | 572 | 579 | 1A,m8 | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-182 | 975 | 981 | 1A | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| miR-190 | 937 | 944 | 1A,m8 | hsa-miR-190 | UGAUAUGUUUGAUAUAUUAGGU |
| miR-363 | 570 | 576 | 1A | hsa-miR-363 | AUUGCACGGUAUCCAUCUGUAA |
| miR-365 | 266 | 272 | m8 | hsa-miR-365 | UAAUGCCCCUAAAAAUCCUUAU |
| miR-369-3p | 251 | 257 | m8 | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-375 | 207 | 213 | 1A | hsa-miR-375 | UUUGUUCGUUCGGCUCGCGUGA |
| miR-450 | 572 | 578 | 1A | hsa-miR-450 | UUUUUGCGAUGUGUUCCUAAUA |
| miR-7 | 681 | 687 | m8 | hsa-miR-7SZ | UGGAAGACUAGUGAUUUUGUUG |
| miR-96 | 975 | 981 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.