Gene Page: VTA1
Summary ?
| GeneID | 51534 |
| Symbol | VTA1 |
| Synonyms | C6orf55|DRG-1|DRG1|HSPC228|LIP5|My012|SBP1 |
| Description | vesicle (multivesicular body) trafficking 1 |
| Reference | MIM:610902|HGNC:HGNC:20954|Ensembl:ENSG00000009844|HPRD:09855| |
| Gene type | protein-coding |
| Map location | 6q24.1 |
| Pascal p-value | 0.274 |
| Sherlock p-value | 0.856 |
| Fetal beta | 0.625 |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10994209 | chr10 | 61877705 | VTA1 | 51534 | 0.05 | trans |
Section II. Transcriptome annotation
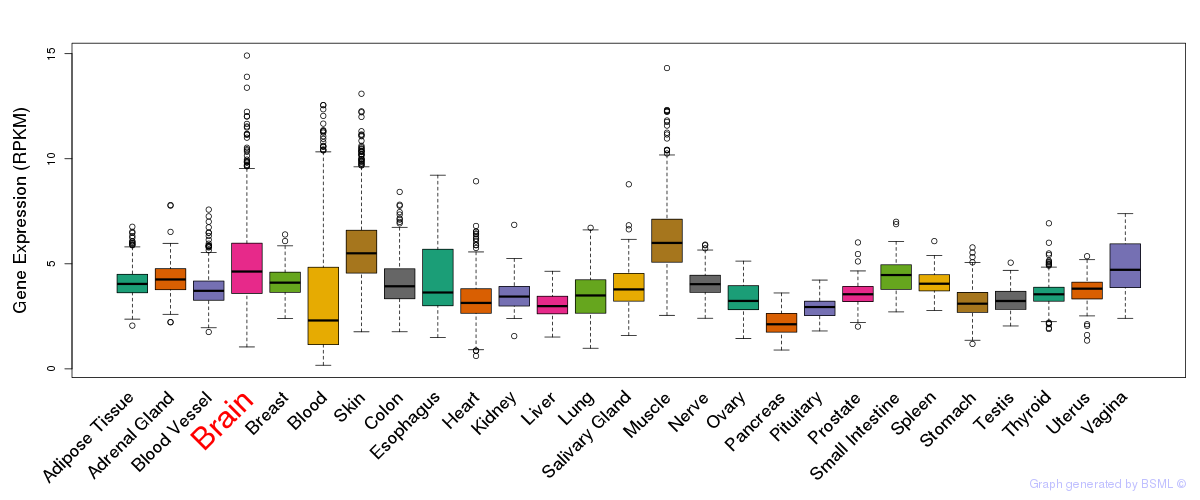
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CBS | HIP4 | cystathionine-beta-synthase | Two-hybrid | BioGRID | 16189514 |
| CDK4 | CMM3 | MGC14458 | PSK-J3 | cyclin-dependent kinase 4 | Two-hybrid | BioGRID | 16189514 |
| DPPA2 | PESCRG1 | developmental pluripotency associated 2 | Two-hybrid | BioGRID | 16189514 |
| KCTD13 | FKSG86 | PDIP1 | POLDIP1 | potassium channel tetramerisation domain containing 13 | Two-hybrid | BioGRID | 16189514 |
| KLHL12 | C3IP1 | DKIR | FLJ27152 | kelch-like 12 (Drosophila) | Two-hybrid | BioGRID | 16189514 |
| LYST | CHS | CHS1 | lysosomal trafficking regulator | Two-hybrid | BioGRID | 11984006 |
| MAGEA11 | MAGE-11 | MAGE11 | MAGEA-11 | MGC10511 | melanoma antigen family A, 11 | Two-hybrid | BioGRID | 16189514 |
| MBIP | - | MAP3K12 binding inhibitory protein 1 | Two-hybrid | BioGRID | 16189514 |
| NBR1 | 1A1-3B | KIAA0049 | M17S2 | MIG19 | neighbor of BRCA1 gene 1 | Two-hybrid | BioGRID | 16189514 |
| NECAB2 | EFCBP2 | N-terminal EF-hand calcium binding protein 2 | Two-hybrid | BioGRID | 16189514 |
| RABAC1 | PRA1 | PRAF1 | YIP3 | Rab acceptor 1 (prenylated) | Two-hybrid | BioGRID | 16189514 |
| RPIA | RPI | ribose 5-phosphate isomerase A | Two-hybrid | BioGRID | 16189514 |
| SSSCA1 | p27 | Sjogren syndrome/scleroderma autoantigen 1 | Two-hybrid | BioGRID | 16189514 |
| ZBTB16 | PLZF | ZNF145 | zinc finger and BTB domain containing 16 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| REACTOME MEMBRANE TRAFFICKING | 129 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | 27 | 15 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| CHANG CORE SERUM RESPONSE UP | 212 | 128 | All SZGR 2.0 genes in this pathway |
| WENDT COHESIN TARGETS UP | 33 | 19 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 | 182 | 102 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |