Gene Page: VIT
Summary ?
| GeneID | 5212 |
| Symbol | VIT |
| Synonyms | - |
| Description | vitrin |
| Reference | HGNC:HGNC:12697|Ensembl:ENSG00000205221|HPRD:18284|Vega:OTTHUMG00000152149 |
| Gene type | protein-coding |
| Map location | 2p22.2 |
| Pascal p-value | 0.006 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| VIT | chr2 | 36970391 | C | T | NM_001177969 NM_001177970 NM_001177971 NM_001177972 NM_053276 | . . . . . | silent silent silent silent silent | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6714112 | chr2 | 36905012 | VIT | 5212 | 0.05 | cis | ||
| rs4670589 | chr2 | 36929220 | VIT | 5212 | 0.08 | cis | ||
| rs644651 | chr1 | 43683365 | VIT | 5212 | 0.18 | trans | ||
| rs1387871 | chr3 | 67343837 | VIT | 5212 | 0.06 | trans | ||
| rs13362722 | chr6 | 6171044 | VIT | 5212 | 0.09 | trans | ||
| rs10993634 | chr9 | 93453379 | VIT | 5212 | 0.13 | trans | ||
| rs10993640 | chr9 | 93467200 | VIT | 5212 | 0.13 | trans | ||
| rs10509166 | chr10 | 63833921 | VIT | 5212 | 0.01 | trans | ||
| rs16906076 | chr11 | 20360967 | VIT | 5212 | 0.02 | trans | ||
| rs5943566 | chrX | 28401493 | VIT | 5212 | 0.05 | trans |
Section II. Transcriptome annotation
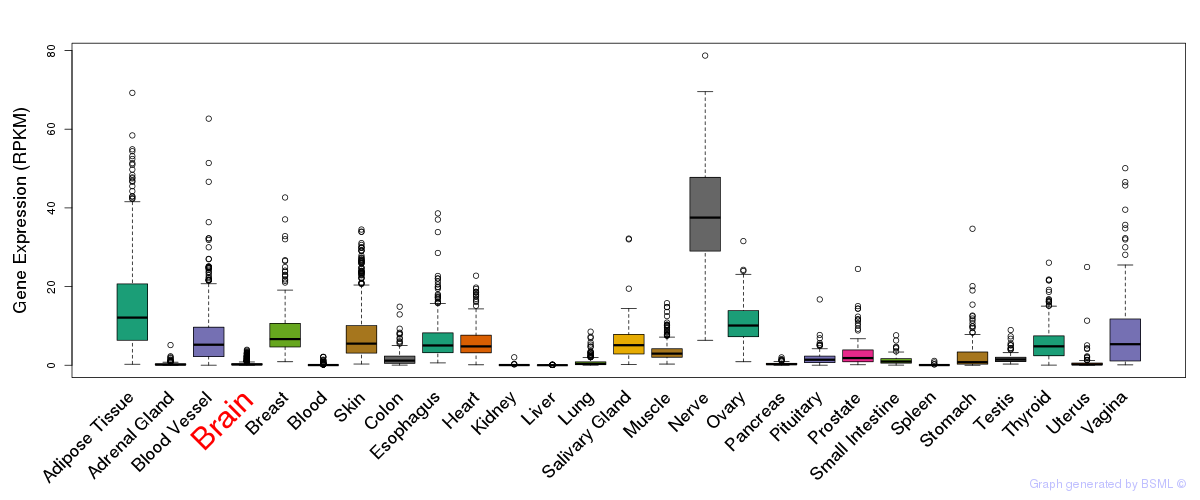
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LIPG | 0.68 | 0.37 |
| LTBP1 | 0.66 | 0.48 |
| DNMBP | 0.66 | 0.49 |
| EEPD1 | 0.65 | 0.47 |
| JUB | 0.64 | 0.45 |
| NOTCH1 | 0.64 | 0.46 |
| TGIF2 | 0.64 | 0.32 |
| MCM3 | 0.64 | 0.45 |
| STON1-GTF2A1L | 0.64 | 0.39 |
| PALLD | 0.64 | 0.38 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CA11 | -0.28 | -0.27 |
| GALNTL5 | -0.28 | -0.29 |
| MT1E | -0.28 | -0.25 |
| MT1X | -0.26 | -0.23 |
| MT1G | -0.25 | -0.29 |
| CISD3 | -0.25 | -0.26 |
| NDUFA4 | -0.25 | -0.24 |
| AC019206.3 | -0.25 | -0.29 |
| SFTPC | -0.25 | -0.23 |
| AF347015.27 | -0.24 | -0.23 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| COLINA TARGETS OF 4EBP1 AND 4EBP2 | 356 | 214 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP | 397 | 206 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| NABA ECM GLYCOPROTEINS | 196 | 99 | All SZGR 2.0 genes in this pathway |
| NABA CORE MATRISOME | 275 | 148 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |