Gene Page: PKLR
Summary ?
| GeneID | 5313 |
| Symbol | PKLR |
| Synonyms | PK1|PKL|PKR|PKRL|RPK |
| Description | pyruvate kinase, liver and RBC |
| Reference | MIM:609712|HGNC:HGNC:9020|Ensembl:ENSG00000143627|HPRD:11841|Vega:OTTHUMG00000035875 |
| Gene type | protein-coding |
| Map location | 1q21 |
| Fetal beta | 0.294 |
| DMG | 1 (# studies) |
| Support | G2Cdb.human_clathrin G2Cdb.human_Synaptosome G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg23109894 | 1 | 155261708 | PKLR | 5.4E-4 | 0.477 | 0.048 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
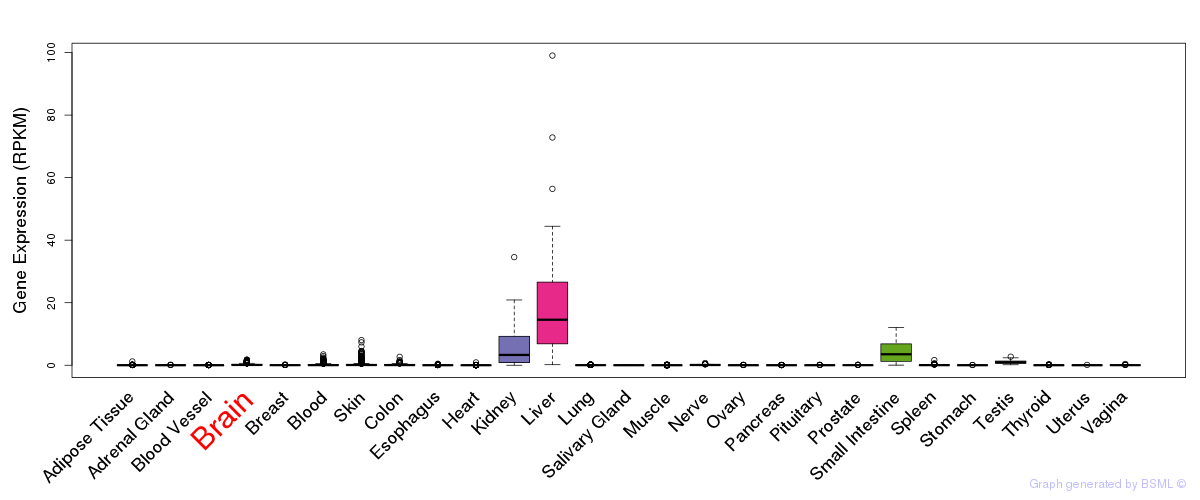
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC130343.3 | 0.57 | 0.53 |
| GEFT | 0.46 | 0.35 |
| FBXL8 | 0.44 | 0.40 |
| TUBA3D | 0.44 | 0.31 |
| POLR2J3 | 0.44 | 0.35 |
| AC131159.1 | 0.43 | 0.33 |
| GBGT1 | 0.42 | 0.37 |
| GPR77 | 0.41 | 0.33 |
| TNFRSF6B | 0.41 | 0.40 |
| ARID5A | 0.41 | 0.30 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TM4SF18 | -0.26 | -0.26 |
| IPMK | -0.21 | -0.11 |
| ZBTB41 | -0.19 | -0.07 |
| FSD1L | -0.19 | -0.07 |
| PABPC4L | -0.19 | -0.14 |
| MYLK2 | -0.19 | -0.18 |
| RSG17 | -0.19 | -0.08 |
| NRAS | -0.19 | -0.08 |
| ITM2A | -0.18 | -0.23 |
| HSPA13 | -0.18 | -0.12 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000287 | magnesium ion binding | IEA | - | |
| GO:0004743 | pyruvate kinase activity | NAS | 1445295 | |
| GO:0004743 | pyruvate kinase activity | TAS | 3126495 | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0030955 | potassium ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006096 | glycolysis | IEA | - | |
| GO:0006096 | glycolysis | NAS | - | |
| GO:0051707 | response to other organism | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005575 | cellular_component | ND | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG GLYCOLYSIS GLUCONEOGENESIS | 62 | 41 | All SZGR 2.0 genes in this pathway |
| KEGG PURINE METABOLISM | 159 | 96 | All SZGR 2.0 genes in this pathway |
| KEGG PYRUVATE METABOLISM | 40 | 26 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG TYPE II DIABETES MELLITUS | 47 | 41 | All SZGR 2.0 genes in this pathway |
| KEGG MATURITY ONSET DIABETES OF THE YOUNG | 25 | 19 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GLYCOLYSIS PATHWAY | 10 | 9 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CHREBP2 PATHWAY | 42 | 35 | All SZGR 2.0 genes in this pathway |
| ST INTEGRIN SIGNALING PATHWAY | 82 | 62 | All SZGR 2.0 genes in this pathway |
| PID HNF3B PATHWAY | 45 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF BETA CELL DEVELOPMENT | 30 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | 20 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOLYSIS | 29 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRATION OF ENERGY METABOLISM | 120 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCOSE METABOLISM | 69 | 42 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING VIA SMAD4 DN | 66 | 38 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 1Q21 AMPLICON | 38 | 18 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR DN | 148 | 102 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL UP | 185 | 112 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE D7 DN | 40 | 22 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SURVIVAL DN | 113 | 76 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF LCP WITH H3K27ME3 | 70 | 35 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS LCP WITH H3K4ME3 | 174 | 100 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES LCP WITH H3K4ME3 | 142 | 80 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 2 | 86 | 50 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS DN | 109 | 71 | All SZGR 2.0 genes in this pathway |