Gene Page: SYTL2
Summary ?
| GeneID | 54843 |
| Symbol | SYTL2 |
| Synonyms | CHR11SYT|EXO4|PPP1R151|SGA72M|SLP2|SLP2A |
| Description | synaptotagmin like 2 |
| Reference | MIM:612880|HGNC:HGNC:15585|Ensembl:ENSG00000137501|HPRD:10260|Vega:OTTHUMG00000166977 |
| Gene type | protein-coding |
| Map location | 11q14 |
| Pascal p-value | 0.084 |
| Sherlock p-value | 0.936 |
| DEG p-value | DEG:Zhao_2015:p=4.51e-04:q=0.0926 |
| Fetal beta | -2.918 |
| eGene | Myers' cis & trans |
| Support | EXOCYTOSIS |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Zhao_2015 | RNA Sequencing analysis | Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs639688 | chr11 | 85470892 | SYTL2 | 54843 | 0.03 | cis | ||
| rs17817179 | chr11 | 85525568 | SYTL2 | 54843 | 0.12 | cis | ||
| rs12335203 | chr8 | 73953618 | SYTL2 | 54843 | 0.09 | trans | ||
| rs7186114 | chr16 | 66839221 | SYTL2 | 54843 | 0.2 | trans |
Section II. Transcriptome annotation
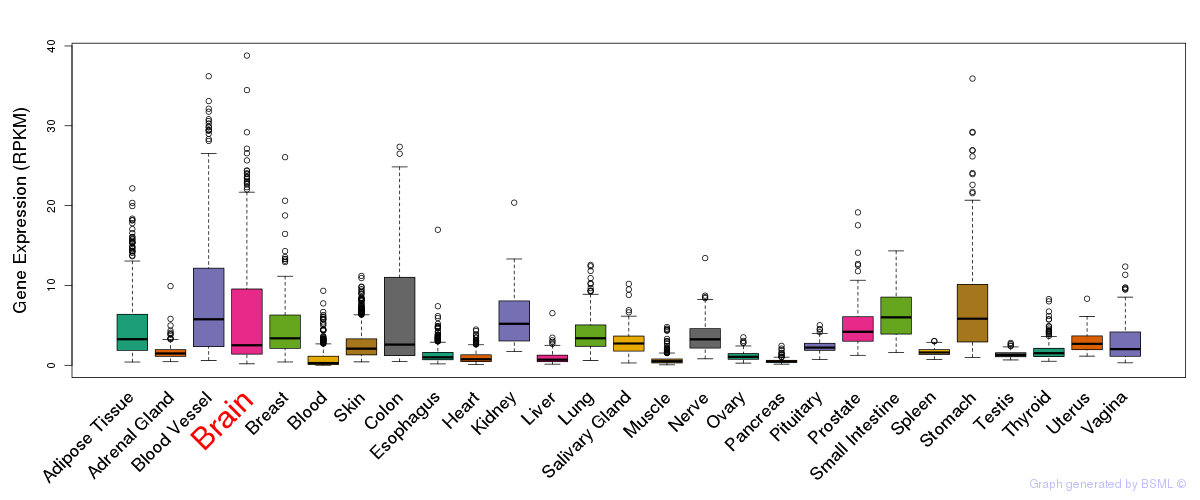
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RMI1 | 0.89 | 0.86 |
| RECQL | 0.88 | 0.84 |
| SLMO2 | 0.88 | 0.86 |
| COPB1 | 0.87 | 0.86 |
| ASF1A | 0.87 | 0.81 |
| CPSF3 | 0.87 | 0.84 |
| ZNF561 | 0.86 | 0.87 |
| KIAA1712 | 0.86 | 0.84 |
| LIN9 | 0.86 | 0.83 |
| AP002373.1 | 0.86 | 0.80 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.61 | -0.72 |
| AF347015.8 | -0.60 | -0.72 |
| AF347015.31 | -0.60 | -0.71 |
| MT-CYB | -0.60 | -0.71 |
| AF347015.27 | -0.60 | -0.70 |
| AF347015.15 | -0.58 | -0.72 |
| AF347015.33 | -0.58 | -0.69 |
| AF347015.2 | -0.57 | -0.69 |
| IFI27 | -0.56 | -0.69 |
| AF347015.21 | -0.55 | -0.69 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0042043 | neurexin binding | ISS | Synap (GO term level: 4) | - |
| GO:0005215 | transporter activity | IEA | - | |
| GO:0017137 | Rab GTPase binding | IEA | - | |
| GO:0031177 | phosphopantetheine binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006810 | transport | IEA | - | |
| GO:0006886 | intracellular protein transport | IEA | - | |
| GO:0016192 | vesicle-mediated transport | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008021 | synaptic vesicle | IEA | Synap, Neurotransmitter (GO term level: 12) | - |
| GO:0012505 | endomembrane system | IEA | - | |
| GO:0005624 | membrane fraction | ISS | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0019897 | extrinsic to plasma membrane | ISS | - | |
| GO:0042470 | melanosome | ISS | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN | 391 | 222 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| BASAKI YBX1 TARGETS DN | 384 | 230 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS DN | 352 | 225 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN | 232 | 139 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP UP | 265 | 158 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER GRADES 1 2 UP | 137 | 84 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA DN | 77 | 52 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| YORDY RECIPROCAL REGULATION BY ETS1 AND SP100 DN | 87 | 48 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED MODERATELY VS POORLY UP | 121 | 71 | All SZGR 2.0 genes in this pathway |
| LIN SILENCED BY TUMOR MICROENVIRONMENT | 108 | 73 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 11Q12 Q14 AMPLICON | 158 | 93 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY MUTATED AND AMPLIFIED IN BREAST CANCER | 94 | 60 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| TRAYNOR RETT SYNDROM UP | 45 | 33 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD UP | 222 | 139 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P3 | 160 | 103 | All SZGR 2.0 genes in this pathway |
| GAVIN PDE3B TARGETS | 22 | 15 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| OSADA ASCL1 TARGETS UP | 46 | 30 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| LEE RECENT THYMIC EMIGRANT | 227 | 128 | All SZGR 2.0 genes in this pathway |
| GYORFFY MITOXANTRONE RESISTANCE | 57 | 35 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR DN | 505 | 328 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 DN | 88 | 61 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 DN | 82 | 51 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| KATSANOU ELAVL1 TARGETS UP | 169 | 105 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-384 | 234 | 241 | 1A,m8 | hsa-miR-384 | AUUCCUAGAAAUUGUUCAUA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.