Gene Page: ZNF821
Summary ?
| GeneID | 55565 |
| Symbol | ZNF821 |
| Synonyms | - |
| Description | zinc finger protein 821 |
| Reference | HGNC:HGNC:28043|Ensembl:ENSG00000102984|HPRD:14244|Vega:OTTHUMG00000176874 |
| Gene type | protein-coding |
| Map location | 16q22.2 |
| Pascal p-value | 0.212 |
| DEG p-value | DEG:Sanders_2014:DS1_p=-0.152:DS1_beta=0.023100:DS2_p=7.11e-01:DS2_beta=-0.018:DS2_FDR=8.62e-01 |
| Fetal beta | 1.591 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Sanders_2013 | Microarray | Whole-genome gene expression profiles using microarrays on lymphoblastoid cell lines (LCLs) from 413 cases and 446 controls. | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg06464432 | 16 | 71917996 | ZNF821 | 8.01E-9 | -0.008 | 3.83E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6734526 | chr2 | 5616370 | ZNF821 | 55565 | 0.18 | trans | ||
| rs16829545 | chr2 | 151977407 | ZNF821 | 55565 | 3.552E-10 | trans | ||
| rs3845734 | chr2 | 171125572 | ZNF821 | 55565 | 0.1 | trans | ||
| rs7584986 | chr2 | 184111432 | ZNF821 | 55565 | 0.04 | trans | ||
| rs17762315 | chr5 | 76807576 | ZNF821 | 55565 | 6.907E-4 | trans | ||
| rs7729096 | chr5 | 76835927 | ZNF821 | 55565 | 0.04 | trans | ||
| rs7787830 | chr7 | 98797019 | ZNF821 | 55565 | 0.01 | trans | ||
| rs2393316 | chr10 | 59333070 | ZNF821 | 55565 | 0.14 | trans | ||
| rs16955618 | chr15 | 29937543 | ZNF821 | 55565 | 1.489E-11 | trans |
Section II. Transcriptome annotation
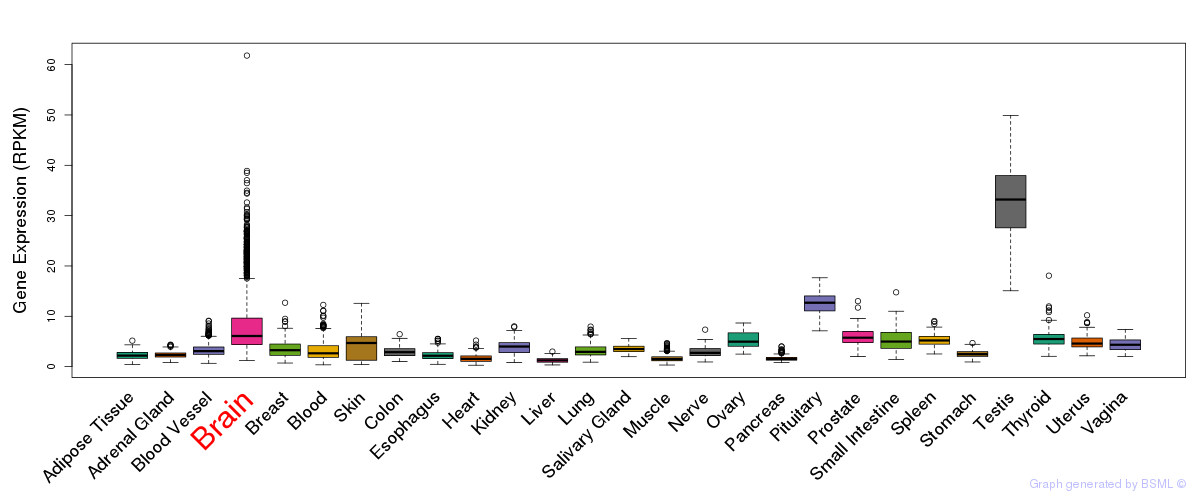
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC112641.2 | 0.83 | 0.88 |
| PITPNM2 | 0.80 | 0.84 |
| DLGAP1 | 0.78 | 0.79 |
| AC021534.2 | 0.78 | 0.85 |
| THRB | 0.78 | 0.80 |
| ARHGEF3 | 0.78 | 0.82 |
| PKNOX2 | 0.77 | 0.80 |
| KCNV1 | 0.77 | 0.84 |
| CAMKK2 | 0.77 | 0.78 |
| IQSEC2 | 0.77 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RAB13 | -0.50 | -0.58 |
| DBI | -0.48 | -0.51 |
| RAB34 | -0.47 | -0.52 |
| MYL12A | -0.46 | -0.52 |
| PECI | -0.45 | -0.51 |
| NKAIN4 | -0.45 | -0.54 |
| AP002478.3 | -0.42 | -0.45 |
| C1orf61 | -0.41 | -0.48 |
| ACAA2 | -0.41 | -0.40 |
| RHOC | -0.41 | -0.46 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP | 178 | 111 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS DN | 135 | 88 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS DN | 242 | 146 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |