Gene Page: CNDP2
Summary ?
| GeneID | 55748 |
| Symbol | CNDP2 |
| Synonyms | CN2|CPGL|HEL-S-13|HsT2298|PEPA |
| Description | CNDP dipeptidase 2 (metallopeptidase M20 family) |
| Reference | MIM:169800|HGNC:HGNC:24437|Ensembl:ENSG00000133313|HPRD:09893|Vega:OTTHUMG00000132853 |
| Gene type | protein-coding |
| Map location | 18q22.3 |
| Pascal p-value | 0.667 |
| Sherlock p-value | 0.957 |
| DMG | 2 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Nucleus accumbens basal ganglia Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
| DNM:Gulsuner_2013 | Whole Exome Sequencing analysis | 155 DNMs identified by exome sequencing of quads or trios of schizophrenia individuals and their parents. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CNDP2 | chr18 | 72180842 | A | C | NM_001168499 NM_018235 | p.180E>A p.264E>A | missense missense | Schizophrenia | DNM:Gulsuner_2013 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01794711 | 18 | 72163198 | CNDP2 | -0.023 | 0.26 | DMG:Nishioka_2013 | |
| cg03669052 | 18 | 72164002 | CNDP2 | 1.26E-8 | -0.007 | 5.11E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs8083001 | 18 | 72165058 | CNDP2 | ENSG00000133313.10 | 7.43746E-7 | 0.03 | 2007 | gtex_brain_ba24 |
| rs3764509 | 18 | 72167124 | CNDP2 | ENSG00000133313.10 | 6.75222E-7 | 0.03 | 4073 | gtex_brain_ba24 |
| rs8084109 | 18 | 72168312 | CNDP2 | ENSG00000133313.10 | 6.70843E-7 | 0.03 | 5261 | gtex_brain_ba24 |
| rs8084410 | 18 | 72168486 | CNDP2 | ENSG00000133313.10 | 6.70843E-7 | 0.03 | 5435 | gtex_brain_ba24 |
| rs7229256 | 18 | 72169129 | CNDP2 | ENSG00000133313.10 | 6.70843E-7 | 0.03 | 6078 | gtex_brain_ba24 |
| rs7241558 | 18 | 72169267 | CNDP2 | ENSG00000133313.10 | 6.70843E-7 | 0.03 | 6216 | gtex_brain_ba24 |
| rs34676212 | 18 | 72170006 | CNDP2 | ENSG00000133313.10 | 6.70843E-7 | 0.03 | 6955 | gtex_brain_ba24 |
| rs4891559 | 18 | 72170397 | CNDP2 | ENSG00000133313.10 | 6.71091E-7 | 0.03 | 7346 | gtex_brain_ba24 |
| rs12971120 | 18 | 72174023 | CNDP2 | ENSG00000133313.10 | 2.66506E-6 | 0.03 | 10972 | gtex_brain_ba24 |
Section II. Transcriptome annotation
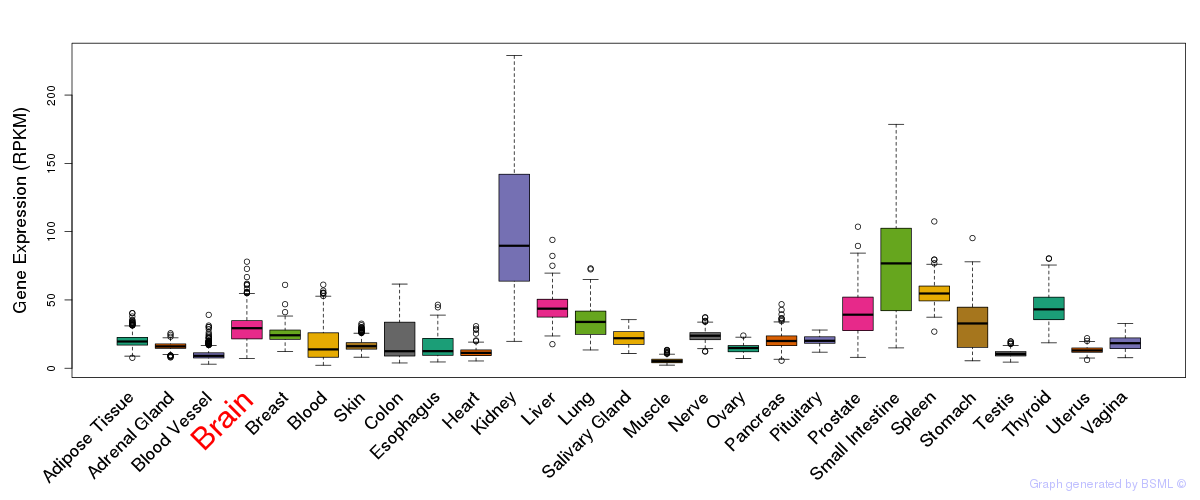
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CHCHD10 | 0.70 | 0.57 |
| KB-1839H6.1 | 0.70 | 0.56 |
| TMEM42 | 0.70 | 0.66 |
| FAM71E1 | 0.69 | 0.65 |
| FAM98C | 0.69 | 0.67 |
| TUSC2 | 0.68 | 0.64 |
| PGP | 0.68 | 0.69 |
| FDX1L | 0.67 | 0.69 |
| HSD11B1L | 0.67 | 0.68 |
| FAM108A4 | 0.67 | 0.64 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EIF5B | -0.38 | -0.47 |
| THOC2 | -0.38 | -0.38 |
| MAP4K4 | -0.34 | -0.33 |
| TOP2B | -0.34 | -0.24 |
| ZEB2 | -0.34 | -0.30 |
| ZNF326 | -0.33 | -0.33 |
| CEP170 | -0.33 | -0.21 |
| RBMX2 | -0.33 | -0.41 |
| AC005035.1 | -0.33 | -0.34 |
| TAF1A | -0.33 | -0.28 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME SULFUR AMINO ACID METABOLISM | 24 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | 200 | 136 | All SZGR 2.0 genes in this pathway |
| REACTOME BIOLOGICAL OXIDATIONS | 139 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUTATHIONE CONJUGATION | 23 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME PHASE II CONJUGATION | 70 | 42 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION DN | 84 | 54 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO UP | 205 | 126 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT UP | 165 | 100 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS DN | 145 | 93 | All SZGR 2.0 genes in this pathway |
| CHUNG BLISTER CYTOTOXICITY UP | 134 | 84 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 5 | 30 | 22 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |