Gene Page: PSEN2
Summary ?
| GeneID | 5664 |
| Symbol | PSEN2 |
| Synonyms | AD3L|AD4|CMD1V|PS2|STM2 |
| Description | presenilin 2 |
| Reference | MIM:600759|HGNC:HGNC:9509|Ensembl:ENSG00000143801|HPRD:02860|Vega:OTTHUMG00000037563 |
| Gene type | protein-coding |
| Map location | 1q42.13 |
| Sherlock p-value | 0.72 |
| Fetal beta | -1.21 |
| eGene | Meta |
| Support | NEUROTROPHIN SIGNALING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0687 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
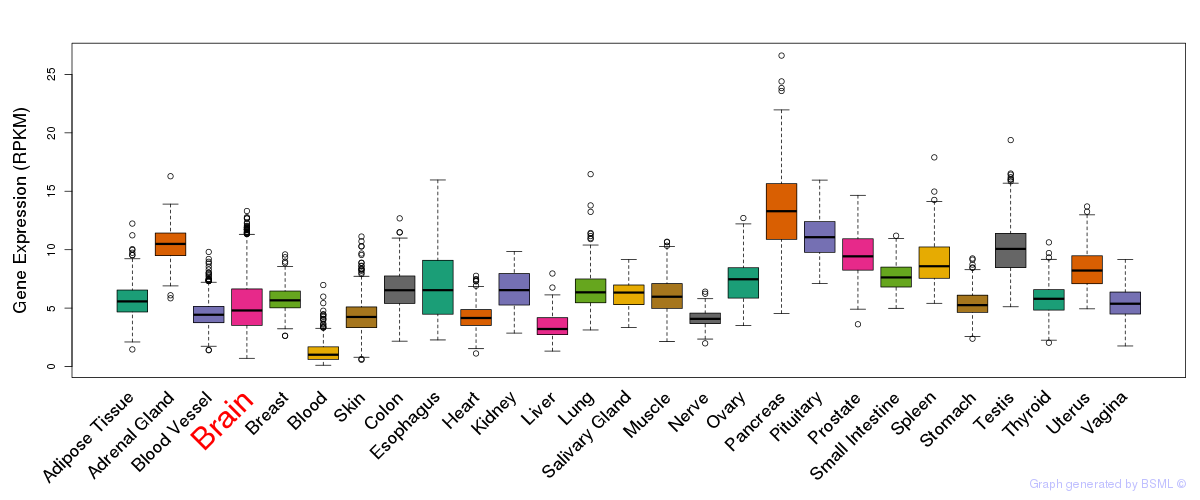
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| IFI35 | 0.95 | 0.94 |
| MT1E | 0.91 | 0.90 |
| RTP4 | 0.90 | 0.84 |
| PSMB9 | 0.90 | 0.93 |
| B2M | 0.89 | 0.90 |
| IFITM1 | 0.89 | 0.88 |
| TAP1 | 0.87 | 0.83 |
| RARRES3 | 0.87 | 0.92 |
| OAS1 | 0.87 | 0.83 |
| OASL | 0.87 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ANKRD17 | -0.68 | -0.79 |
| MCM3AP | -0.68 | -0.81 |
| EIF4ENIF1 | -0.68 | -0.79 |
| MYO18A | -0.68 | -0.72 |
| SFRS14 | -0.68 | -0.81 |
| HEATR5B | -0.68 | -0.82 |
| USP7 | -0.68 | -0.77 |
| CKAP5 | -0.68 | -0.82 |
| PDXDC1 | -0.67 | -0.78 |
| SPTAN1 | -0.67 | -0.80 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 12297508 | |
| GO:0008233 | peptidase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0042987 | amyloid precursor protein catabolic process | TAS | 15274632 | |
| GO:0006509 | membrane protein ectodomain proteolysis | IDA | 15274632 | |
| GO:0007220 | Notch receptor processing | TAS | 15274632 | |
| GO:0008632 | apoptotic program | TAS | 8939861 | |
| GO:0007059 | chromosome segregation | TAS | 9298903 | |
| GO:0016485 | protein processing | IDA | 15274632 | |
| GO:0043085 | positive regulation of catalytic activity | IDA | 15274632 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000139 | Golgi membrane | IEA | - | |
| GO:0000776 | kinetochore | TAS | 9298903 | |
| GO:0005794 | Golgi apparatus | IDA | 15274632 | |
| GO:0005789 | endoplasmic reticulum membrane | IEA | - | |
| GO:0005639 | integral to nuclear inner membrane | TAS | 9298903 | |
| GO:0005783 | endoplasmic reticulum | IDA | 15274632 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | IDA | 15274632 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APBA1 | D9S411E | MINT1 | X11 | X11A | X11ALPHA | amyloid beta (A4) precursor protein-binding, family A, member 1 | Affinity Capture-Western | BioGRID | 12196555 |
| APBA2 | D15S1518E | HsT16821 | LIN-10 | MGC99508 | MGC:14091 | MINT2 | X11L | amyloid beta (A4) precursor protein-binding, family A, member 2 | Affinity Capture-Western | BioGRID | 12196555 |
| APBA3 | MGC:15815 | X11L2 | mint3 | amyloid beta (A4) precursor protein-binding, family A, member 3 | Affinity Capture-Western | BioGRID | 12196555 |
| APH1A | 6530402N02Rik | APH-1A | CGI-78 | anterior pharynx defective 1 homolog A (C. elegans) | - | HPRD,BioGRID | 12297508 |12471034 |
| APH1B | APH-1B | DKFZp564D0372 | PRO1328 | PSFL | TAAV688 | anterior pharynx defective 1 homolog B (C. elegans) | - | HPRD | 12297508 |
| APP | AAA | ABETA | ABPP | AD1 | APPI | CTFgamma | CVAP | PN2 | amyloid beta (A4) precursor protein | PS2 interacts with APP. | BIND | 9223340 |
| BCL2L1 | BCL-XL/S | BCL2L | BCLX | Bcl-X | DKFZp781P2092 | bcl-xL | bcl-xS | BCL2-like 1 | - | HPRD,BioGRID | 10446169 |
| CAPN1 | CANP | CANP1 | CANPL1 | muCANP | muCL | calpain 1, (mu/I) large subunit | - | HPRD,BioGRID | 9852298 |
| CIB1 | CIB | KIP | KIP1 | SIP2-28 | calcium and integrin binding 1 (calmyrin) | - | HPRD,BioGRID | 10366599 |
| CTNND2 | GT24 | NPRAP | catenin (cadherin-associated protein), delta 2 (neural plakophilin-related arm-repeat protein) | - | HPRD | 10037471 |
| DOCK3 | KIAA0299 | MOCA | PBP | dedicator of cytokinesis 3 | - | HPRD | 10854253 |
| FHL2 | AAG11 | DRAL | SLIM3 | four and a half LIM domains 2 | - | HPRD,BioGRID | 11001931 |
| FLNB | ABP-278 | AOI | DKFZp686A1668 | DKFZp686O033 | FH1 | FLN1L | LRS1 | SCT | TABP | TAP | filamin B, beta (actin binding protein 278) | - | HPRD,BioGRID | 9437013 |
| GFAP | FLJ45472 | glial fibrillary acidic protein | Two-hybrid | BioGRID | 12058025 |
| ICAM5 | TLCN | TLN | intercellular adhesion molecule 5, telencephalin | - | HPRD,BioGRID | 11719200 |
| KCNIP3 | CSEN | DREAM | KCHIP3 | MGC18289 | Kv channel interacting protein 3, calsenilin | - | HPRD,BioGRID | 9771752 |
| KCNIP4 | CALP | KCHIP4 | MGC44947 | Kv channel interacting protein 4 | - | HPRD,BioGRID | 11847232 |
| METTL2B | FLJ11350 | FLJ12760 | METL | METTL2 | METTL2A | PSENIP1 | methyltransferase like 2B | - | HPRD,BioGRID | 11738826 |
| NCSTN | APH2 | KIAA0253 | nicastrin | - | HPRD,BioGRID | 10993067 |
| PSEN1 | AD3 | FAD | PS1 | S182 | presenilin 1 | Affinity Capture-Western | BioGRID | 12471034 |
| PSENEN | MDS033 | MSTP064 | PEN-2 | PEN2 | presenilin enhancer 2 homolog (C. elegans) | - | HPRD,BioGRID | 12198112 |12639958 |
| SRI | FLJ26259 | SCN | sorcin | - | HPRD,BioGRID | 10748169 |
| UBQLN1 | DA41 | DSK2 | FLJ90054 | PLIC-1 | XDRP1 | ubiquilin 1 | - | HPRD,BioGRID | 11076969 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NOTCH SIGNALING PATHWAY | 47 | 35 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIVNEF PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| PID LKB1 PATHWAY | 47 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR SIGNALING BY ERBB4 | 38 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | 27 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH4 | 12 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH2 | 12 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH1 | 70 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH3 | 12 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATED PROTEOLYSIS OF P75NTR | 10 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | 15 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | 60 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | 81 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH | 103 | 64 | All SZGR 2.0 genes in this pathway |
| DAVICIONI PAX FOXO1 SIGNATURE IN ARMS UP | 59 | 38 | All SZGR 2.0 genes in this pathway |
| DAVICIONI MOLECULAR ARMS VS ERMS UP | 332 | 228 | All SZGR 2.0 genes in this pathway |
| DAVICIONI TARGETS OF PAX FOXO1 FUSIONS UP | 255 | 177 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL DN | 86 | 59 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA DN | 146 | 94 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HIF1A TARGETS UP | 67 | 40 | All SZGR 2.0 genes in this pathway |
| LIU CDX2 TARGETS UP | 36 | 22 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION DN | 100 | 64 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER DN | 203 | 134 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| SHIN B CELL LYMPHOMA CLUSTER 2 | 30 | 23 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER MYC TGFA DN | 65 | 44 | All SZGR 2.0 genes in this pathway |
| UEDA PERIFERAL CLOCK | 169 | 111 | All SZGR 2.0 genes in this pathway |
| HADDAD T LYMPHOCYTE AND NK PROGENITOR UP | 78 | 56 | All SZGR 2.0 genes in this pathway |
| ABRAHAM ALPC VS MULTIPLE MYELOMA DN | 19 | 14 | All SZGR 2.0 genes in this pathway |
| TAKAO RESPONSE TO UVB RADIATION DN | 98 | 67 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| RAMASWAMY METASTASIS UP | 66 | 43 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 6 | 189 | 112 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P3 | 160 | 103 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF UP | 418 | 282 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN | 245 | 150 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE DN | 72 | 47 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-183 | 74 | 81 | 1A,m8 | hsa-miR-183 | UAUGGCACUGGUAGAAUUCACUG |
| miR-30-5p | 121 | 127 | m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.