Gene Page: JAM2
Summary ?
| GeneID | 58494 |
| Symbol | JAM2 |
| Synonyms | C21orf43|CD322|JAM-B|JAMB|PRO245|VE-JAM|VEJAM |
| Description | junctional adhesion molecule 2 |
| Reference | MIM:606870|HGNC:HGNC:14686|Ensembl:ENSG00000154721|HPRD:06041|Vega:OTTHUMG00000078441 |
| Gene type | protein-coding |
| Map location | 21q21.2 |
| Pascal p-value | 0.082 |
| Sherlock p-value | 0.694 |
| Fetal beta | -0.33 |
| DMG | 2 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg22627390 | 21 | 27012066 | JAM2 | 2.536E-4 | -0.224 | 0.037 | DMG:Wockner_2014 |
| cg03382304 | 21 | 27012176 | JAM2 | -0.02 | 0.26 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17131472 | chr1 | 91962966 | JAM2 | 58494 | 0.06 | trans | ||
| rs1691463 | chr3 | 84914366 | JAM2 | 58494 | 0.03 | trans | ||
| rs2922180 | chr3 | 125280472 | JAM2 | 58494 | 0.17 | trans | ||
| rs2976809 | chr3 | 125451738 | JAM2 | 58494 | 0.04 | trans | ||
| rs10050805 | chr5 | 30805352 | JAM2 | 58494 | 0.08 | trans | ||
| rs17135501 | chr6 | 2704474 | JAM2 | 58494 | 2.549E-5 | trans | ||
| rs17135521 | chr6 | 2707631 | JAM2 | 58494 | 2.549E-5 | trans | ||
| snp_a-1787926 | 0 | JAM2 | 58494 | 2.549E-5 | trans | |||
| rs6455226 | chr6 | 67930295 | JAM2 | 58494 | 0.01 | trans | ||
| rs7841407 | chr8 | 9243427 | JAM2 | 58494 | 0.1 | trans | ||
| rs7812588 | chr8 | 136803829 | JAM2 | 58494 | 0.18 | trans | ||
| rs4514358 | chr10 | 43861923 | JAM2 | 58494 | 0.19 | trans | ||
| rs1159612 | chr10 | 85329057 | JAM2 | 58494 | 0.16 | trans | ||
| rs10492573 | chr13 | 43890007 | JAM2 | 58494 | 0.18 | trans | ||
| rs9891406 | chr17 | 34320983 | JAM2 | 58494 | 0.04 | trans | ||
| rs6095741 | chr20 | 48666589 | JAM2 | 58494 | 0.01 | trans | ||
| rs1545676 | chrX | 93530472 | JAM2 | 58494 | 0.03 | trans |
Section II. Transcriptome annotation
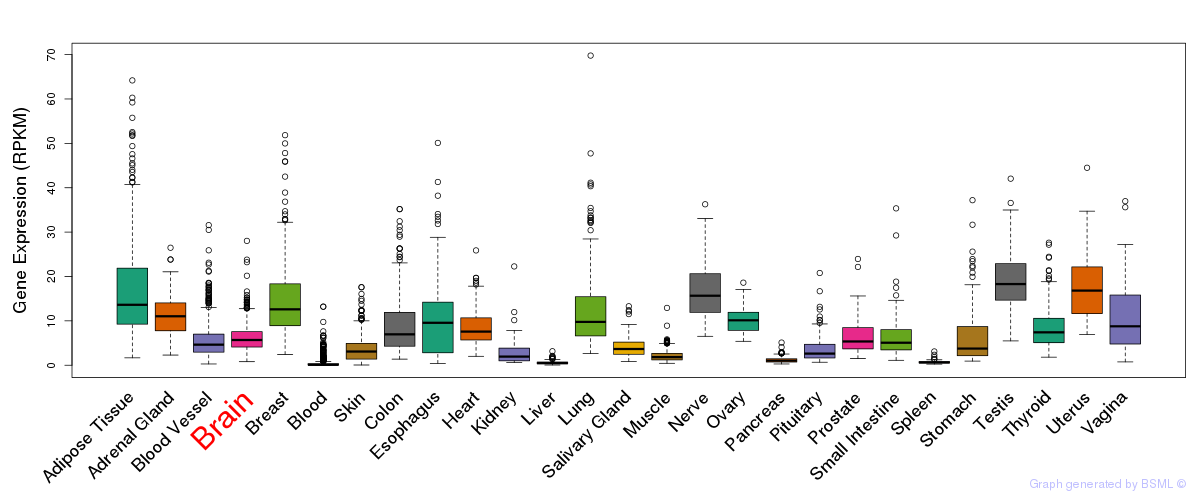
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NDUFB5 | 0.86 | 0.79 |
| C2orf64 | 0.86 | 0.75 |
| NDUFA4 | 0.85 | 0.75 |
| SDHB | 0.84 | 0.78 |
| NDUFA9 | 0.84 | 0.75 |
| COX4I1 | 0.84 | 0.76 |
| AC025647.1 | 0.84 | 0.74 |
| MRPS33 | 0.84 | 0.72 |
| GSTO1 | 0.82 | 0.68 |
| APOA1BP | 0.82 | 0.73 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DMPK | -0.43 | -0.50 |
| AC005921.3 | -0.43 | -0.59 |
| SMTN | -0.42 | -0.55 |
| IL3RA | -0.42 | -0.60 |
| AC010300.1 | -0.41 | -0.52 |
| TNKS1BP1 | -0.41 | -0.38 |
| BAT2D1 | -0.40 | -0.33 |
| PRX | -0.40 | -0.45 |
| UPF3A | -0.40 | -0.48 |
| HDAC7 | -0.40 | -0.50 |
Section III. Gene Ontology annotation
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0016337 | cell-cell adhesion | NAS | 10779521 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005923 | tight junction | IEA | Brain (GO term level: 10) | - |
| GO:0005886 | plasma membrane | EXP | 17525755 | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | NAS | 10779521 | |
| GO:0030054 | cell junction | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL ADHESION MOLECULES CAMS | 134 | 93 | All SZGR 2.0 genes in this pathway |
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| KEGG LEUKOCYTE TRANSENDOTHELIAL MIGRATION | 118 | 78 | All SZGR 2.0 genes in this pathway |
| KEGG EPITHELIAL CELL SIGNALING IN HELICOBACTER PYLORI INFECTION | 68 | 44 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN1 PATHWAY | 66 | 44 | All SZGR 2.0 genes in this pathway |
| PID AMB2 NEUTROPHILS PATHWAY | 41 | 32 | All SZGR 2.0 genes in this pathway |
| PID INTEGRIN A4B1 PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL SURFACE INTERACTIONS AT THE VASCULAR WALL | 91 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME INTEGRIN CELL SURFACE INTERACTIONS | 79 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA DN | 116 | 79 | All SZGR 2.0 genes in this pathway |
| VECCHI GASTRIC CANCER EARLY DN | 367 | 220 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA DN | 291 | 176 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS A UP | 191 | 128 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM DN | 164 | 111 | All SZGR 2.0 genes in this pathway |
| CERVERA SDHB TARGETS 1 UP | 118 | 66 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL LONG TERM | 302 | 191 | All SZGR 2.0 genes in this pathway |
| CHEN LVAD SUPPORT OF FAILING HEART DN | 42 | 32 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| CLASPER LYMPHATIC VESSELS DURING METASTASIS DN | 36 | 23 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL DN | 216 | 143 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING | 243 | 155 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS UP | 112 | 65 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| YANG BCL3 TARGETS UP | 364 | 236 | All SZGR 2.0 genes in this pathway |
| WANG MLL TARGETS | 289 | 188 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 | 227 | 149 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA NS UP | 162 | 103 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 2 TRANSIENTLY INDUCED BY EGF | 51 | 29 | All SZGR 2.0 genes in this pathway |