Gene Page: RALA
Summary ?
| GeneID | 5898 |
| Symbol | RALA |
| Synonyms | RAL |
| Description | v-ral simian leukemia viral oncogene homolog A (ras related) |
| Reference | MIM:179550|HGNC:HGNC:9839|Ensembl:ENSG00000006451|HPRD:01549|Vega:OTTHUMG00000128775 |
| Gene type | protein-coding |
| Map location | 7p15-p13 |
| Pascal p-value | 0.374 |
| Sherlock p-value | 0.492 |
| Fetal beta | 1.098 |
| DMG | 1 (# studies) |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0309 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg11610702 | 7 | 39773227 | RALA | 1.33E-11 | -0.036 | 2.84E-7 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
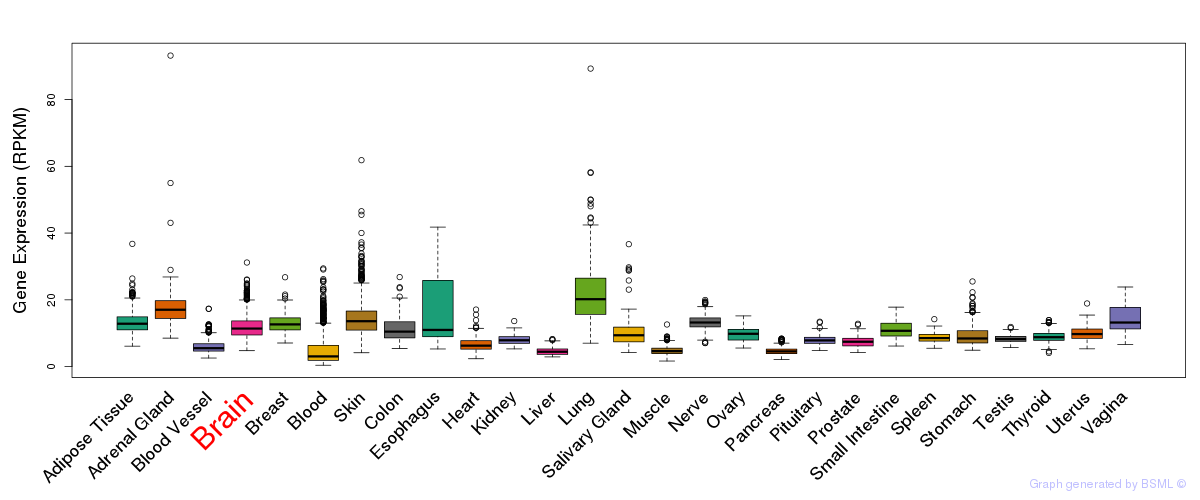
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA1009 | 0.93 | 0.90 |
| KIAA1731 | 0.93 | 0.91 |
| C21orf66 | 0.91 | 0.90 |
| ZNF841 | 0.90 | 0.91 |
| ZFC3H1 | 0.90 | 0.92 |
| CSPP1 | 0.90 | 0.91 |
| RBM6 | 0.90 | 0.88 |
| CEP290 | 0.90 | 0.87 |
| CENPJ | 0.90 | 0.89 |
| ZNF471 | 0.90 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.65 | -0.77 |
| IFI27 | -0.64 | -0.76 |
| ENHO | -0.63 | -0.80 |
| HIGD1B | -0.63 | -0.77 |
| MT-CO2 | -0.63 | -0.75 |
| AF347015.27 | -0.61 | -0.71 |
| TSC22D4 | -0.61 | -0.69 |
| FXYD1 | -0.61 | -0.72 |
| RAMP1 | -0.61 | -0.70 |
| HLA-F | -0.61 | -0.65 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 7673236 | |
| GO:0005525 | GTP binding | TAS | 2550440 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007265 | Ras protein signal transduction | EXP | 11520933 | |
| GO:0006935 | chemotaxis | TAS | 10848592 | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARF1 | - | ADP-ribosylation factor 1 | Reconstituted Complex | BioGRID | 9688545 |
| ARF1 | - | ADP-ribosylation factor 1 | - | HPRD | 12509462 |
| ARF6 | DKFZp564M0264 | ADP-ribosylation factor 6 | Affinity Capture-Western | BioGRID | 12509462 |
| CNKSR2 | CNK2 | KIAA0902 | KSR2 | connector enhancer of kinase suppressor of Ras 2 | - | HPRD,BioGRID | 14597674 |
| CSDA | CSDA1 | DBPA | ZONAB | cold shock domain protein A | RalA interacts with ZONAB. This interaction was modeled on a demonstrated interaction between human or canine RalA and canine ZONAB. | BIND | 15592429 |
| EXOC1 | BM-102 | FLJ10893 | SEC3 | SEC3L1 | SEC3P | exocyst complex component 1 | - | HPRD | 11406615 |
| EXOC2 | FLJ11026 | SEC5 | SEC5L1 | Sec5p | exocyst complex component 2 | - | HPRD,BioGRID | 12839989 |
| EXOC2 | FLJ11026 | SEC5 | SEC5L1 | Sec5p | exocyst complex component 2 | - | HPRD | 11744922 |12624092 |14525976 |
| EXOC2 | FLJ11026 | SEC5 | SEC5L1 | Sec5p | exocyst complex component 2 | RalA interacts with Sec5. This interaction was modelled on a demonstrated interaction between human RalA and Sec5 from an unspecified species. | BIND | 15920473 |
| EXOC4 | MGC27170 | REC8 | SEC8 | SEC8L1 | Sec8p | exocyst complex component 4 | - | HPRD | 11406615 |
| EXOC7 | 2-5-3p | DKFZp686J04253 | EX070 | EXO70 | EXOC1 | Exo70p | FLJ40965 | FLJ46415 | YJL085W | exocyst complex component 7 | - | HPRD | 11406615 |
| EXOC8 | EXO84 | Exo84p | SEC84 | exocyst complex component 8 | - | HPRD,BioGRID | 14525976 |
| FLNA | ABP-280 | ABPX | DKFZp434P031 | FLN | FLN1 | FMD | MNS | NHBP | OPD | OPD1 | OPD2 | filamin A, alpha (actin binding protein 280) | - | HPRD,BioGRID | 10051605 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | Two-hybrid | BioGRID | 11786539 |
| LRPAP1 | A2MRAP | A2RAP | HBP44 | MGC138272 | MRAP | RAP | low density lipoprotein receptor-related protein associated protein 1 | Reconstituted Complex | BioGRID | 11167825 |
| PLD1 | - | phospholipase D1, phosphatidylcholine-specific | - | HPRD,BioGRID | 9207251 |
| RAB8B | FLJ38125 | RAB8B, member RAS oncogene family | - | HPRD | 10889189 |
| RALBP1 | RIP1 | RLIP1 | RLIP76 | ralA binding protein 1 | - | HPRD,BioGRID | 7673236 |
| RALBP1 | RIP1 | RLIP1 | RLIP76 | ralA binding protein 1 | RalA interacts with RalBP1. | BIND | 15592429 |
| RALBP1 | RIP1 | RLIP1 | RLIP76 | ralA binding protein 1 | RalA interacts with RLIP76. | BIND | 7673236 |
| RALGPS1 | KIAA0351 | RALGEF2 | RALGPS1A | Ral GEF with PH domain and SH3 binding motif 1 | - | HPRD,BioGRID | 10889189 |
| RAP1A | KREV-1 | KREV1 | RAP1 | SMGP21 | RAP1A, member of RAS oncogene family | Two-hybrid | BioGRID | 11786539 |
| RAP2A | K-REV | KREV | RAP2 | RbBP-30 | RAP2A, member of RAS oncogene family | Two-hybrid | BioGRID | 11786539 |
| RAP2B | MGC20484 | RAP2B, member of RAS oncogene family | Two-hybrid | BioGRID | 11786539 |
| RGL2 | HKE1.5 | KE1.5 | RAB2L | ral guanine nucleotide dissociation stimulator-like 2 | Two-hybrid | BioGRID | 8939933 |
| RGL4 | MGC119678 | MGC119680 | Rgr | ral guanine nucleotide dissociation stimulator-like 4 | - | HPRD,BioGRID | 12874025 |
| TSC2 | FLJ43106 | LAM | TSC4 | tuberous sclerosis 2 | Affinity Capture-Western | BioGRID | 12842888 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG PANCREATIC CANCER | 70 | 56 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RAS PATHWAY | 23 | 20 | All SZGR 2.0 genes in this pathway |
| ST INTEGRIN SIGNALING PATHWAY | 82 | 62 | All SZGR 2.0 genes in this pathway |
| PID ARF6 TRAFFICKING PATHWAY | 49 | 34 | All SZGR 2.0 genes in this pathway |
| PID P38 ALPHA BETA PATHWAY | 31 | 25 | All SZGR 2.0 genes in this pathway |
| PID FOXO PATHWAY | 49 | 43 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 DOWNSTREAM PATHWAY | 105 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING TO RAS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING TO ERKS | 36 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME P38MAPK EVENTS | 13 | 12 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS TOP50 UP | 38 | 27 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS RED DN | 25 | 19 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP UP | 265 | 158 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER GRADES 1 2 UP | 137 | 84 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| CHOI ATL STAGE PREDICTOR | 44 | 24 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE AUGMENTED BY MYC | 108 | 74 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| OXFORD RALA TARGETS DN | 10 | 8 | All SZGR 2.0 genes in this pathway |
| ASTON MAJOR DEPRESSIVE DISORDER DN | 160 | 110 | All SZGR 2.0 genes in this pathway |
| BROCKE APOPTOSIS REVERSED BY IL6 | 144 | 98 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| STOSSI RESPONSE TO ESTRADIOL | 50 | 35 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS | 108 | 71 | All SZGR 2.0 genes in this pathway |
| MCCLUNG CREB1 TARGETS UP | 100 | 72 | All SZGR 2.0 genes in this pathway |
| WESTON VEGFA TARGETS 3HR | 74 | 47 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP | 388 | 234 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| CHUNG BLISTER CYTOTOXICITY UP | 134 | 84 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| LEE LIVER CANCER SURVIVAL DN | 175 | 103 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS | 535 | 325 | All SZGR 2.0 genes in this pathway |
| CHANDRAN METASTASIS UP | 221 | 135 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 10 | 69 | 38 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| VANOEVELEN MYOGENESIS SIN3A TARGETS | 220 | 133 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL UP | 146 | 75 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 1462 | 1468 | 1A | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-124.1 | 594 | 601 | 1A,m8 | hsa-miR-124a | UUAAGGCACGCGGUGAAUGCCA |
| miR-124/506 | 594 | 600 | 1A | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-153 | 452 | 458 | 1A | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-181 | 238 | 245 | 1A,m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-183 | 772 | 779 | 1A,m8 | hsa-miR-183 | UAUGGCACUGGUAGAAUUCACUG |
| miR-221/222 | 241 | 247 | m8 | hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
| miR-24 | 1838 | 1844 | 1A | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-27 | 519 | 525 | 1A | hsa-miR-27abrain | UUCACAGUGGCUAAGUUCCGC |
| hsa-miR-27bbrain | UUCACAGUGGCUAAGUUCUGC | ||||
| miR-29 | 1760 | 1766 | 1A | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-30-3p | 79 | 85 | m8 | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-329 | 638 | 644 | m8 | hsa-miR-329brain | AACACACCUGGUUAACCUCUUU |
| miR-34b | 389 | 395 | m8 | hsa-miR-34b | UAGGCAGUGUCAUUAGCUGAUUG |
| miR-369-3p | 341 | 347 | 1A | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 341 | 348 | 1A,m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-376 | 1764 | 1770 | 1A | hsa-miR-376a | AUCAUAGAGGAAAAUCCACGU |
| hsa-miR-376b | AUCAUAGAGGAAAAUCCAUGUU | ||||
| miR-448 | 452 | 458 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.