Gene Page: RASA1
Summary ?
| GeneID | 5921 |
| Symbol | RASA1 |
| Synonyms | CM-AVM|CMAVM|GAP|PKWS|RASA|RASGAP|p120|p120GAP|p120RASGAP |
| Description | RAS p21 protein activator 1 |
| Reference | MIM:139150|HGNC:HGNC:9871|Ensembl:ENSG00000145715|HPRD:00745|Vega:OTTHUMG00000162605 |
| Gene type | protein-coding |
| Map location | 5q13.3 |
| Pascal p-value | 0.64 |
| Sherlock p-value | 0.678 |
| Fetal beta | 0.423 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 2 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.2213 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09490071 | 5 | 86564760 | RASA1 | 2.7E-9 | -0.022 | 1.95E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
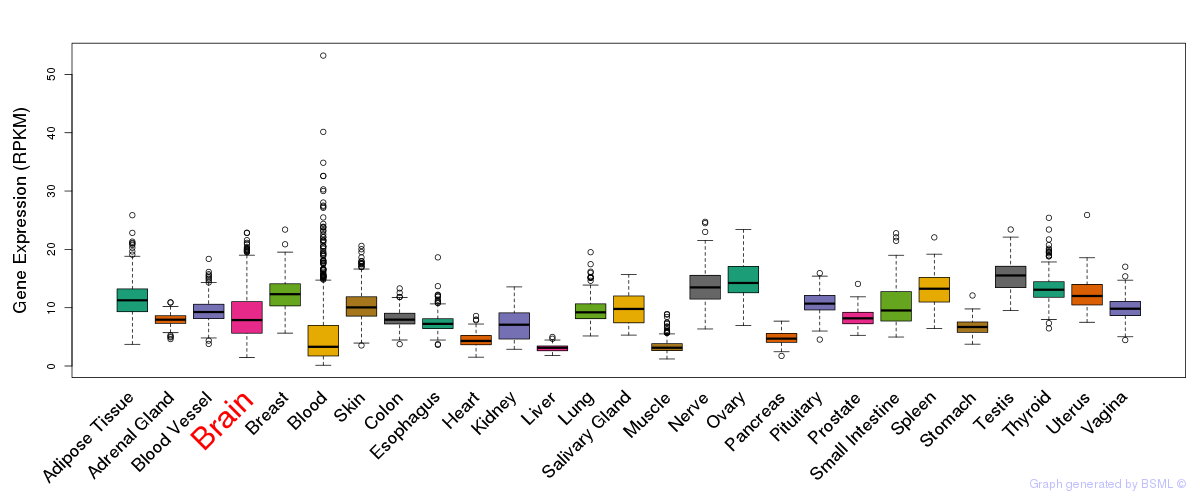
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005102 | receptor binding | IPI | Neurotransmitter (GO term level: 4) | 2157284 |2176151 |
| GO:0005099 | Ras GTPase activator activity | TAS | 2188736 | |
| GO:0019870 | potassium channel inhibitor activity | NAS | 1553544 | |
| GO:0051020 | GTPase binding | IPI | 2122974 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0043524 | negative regulation of neuron apoptosis | ISS | neuron (GO term level: 9) | - |
| GO:0001570 | vasculogenesis | ISS | - | |
| GO:0000910 | cytokinesis | ISS | - | |
| GO:0001953 | negative regulation of cell-matrix adhesion | IDA | 8344248 | |
| GO:0007242 | intracellular signaling cascade | NAS | 1581965 | |
| GO:0008360 | regulation of cell shape | NAS | 9113414 | |
| GO:0009790 | embryonic development | ISS | - | |
| GO:0051056 | regulation of small GTPase mediated signal transduction | IEA | - | |
| GO:0051252 | regulation of RNA metabolic process | NAS | 9113414 | |
| GO:0030833 | regulation of actin filament polymerization | IDA | 8344248 | |
| GO:0045768 | positive regulation of anti-apoptosis | IDA | 15542850 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001726 | ruffle | IEA | - | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005737 | cytoplasm | NAS | 2821624 | |
| GO:0005886 | plasma membrane | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | c-Abl interacts with GAP. | BIND | 1383690 |
| ALK | CD246 | Ki-1 | TFG/ALK | anaplastic lymphoma receptor tyrosine kinase | - | HPRD | 9174053 |
| ANXA6 | ANX6 | CBP68 | annexin A6 | - | HPRD,BioGRID | 8798684 |10571081 |
| ANXA6 | ANX6 | CBP68 | annexin A6 | Annexin VI interacts with GAP. | BIND | 10571081 |
| ARHGAP1 | CDC42GAP | RHOGAP | RHOGAP1 | p50rhoGAP | Rho GTPase activating protein 1 | - | HPRD | 9034330 |
| ARHGAP5 | GFI2 | RhoGAP5 | p190-B | Rho GTPase activating protein 5 | GAP interacts with p190. This interaction was modeled on a demonstrated interaction between human GAP and rat p190. | BIND | 1689011 |8344248 |10769036 |
| ARHGAP5 | GFI2 | RhoGAP5 | p190-B | Rho GTPase activating protein 5 | - | HPRD | 8537347 |
| AURKA | AIK | ARK1 | AURA | AURORA2 | BTAK | MGC34538 | STK15 | STK6 | STK7 | aurora kinase A | RasGAP interacts with HsAIRK-1. | BIND | 11976319 |
| AURKB | AIK2 | AIM-1 | AIM1 | ARK2 | AurB | IPL1 | STK12 | STK5 | aurora kinase B | - | HPRD | 11976319 |
| AURKB | AIK2 | AIM-1 | AIM1 | ARK2 | AurB | IPL1 | STK12 | STK5 | aurora kinase B | RasGAP interacts with HsAIRK-2 in the RasGAP/HsAIRK-2/survivin complex. | BIND | 11976319 |
| BCR | ALL | BCR-ABL1 | BCR1 | CML | D22S11 | D22S662 | FLJ16453 | PHL | breakpoint cluster region | BCR interacts with GAP. This interaction was modeled on a demonstrated interaction between BCR from an unspecified source and human GAP. | BIND | 1383690 |
| BIRC5 | API4 | EPR-1 | baculoviral IAP repeat-containing 5 | - | HPRD | 11976319 |
| CAV2 | CAV | MGC12294 | caveolin 2 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12091389 |
| CD5 | LEU1 | T1 | CD5 molecule | - | HPRD,BioGRID | 9603468 |
| CSF1R | C-FMS | CD115 | CSFR | FIM2 | FMS | colony stimulating factor 1 receptor | - | HPRD,BioGRID | 2172781 |
| CSK | MGC117393 | c-src tyrosine kinase | - | HPRD,BioGRID | 7544435 |
| DNAJA3 | FLJ45758 | TID1 | hTid-1 | DnaJ (Hsp40) homolog, subfamily A, member 3 | - | HPRD,BioGRID | 11116152 |
| DOK1 | MGC117395 | MGC138860 | P62DOK | docking protein 1, 62kDa (downstream of tyrosine kinase 1) | - | HPRD,BioGRID | 9008161 |
| DOK1 | MGC117395 | MGC138860 | P62DOK | docking protein 1, 62kDa (downstream of tyrosine kinase 1) | DOK1 interacts with RASGAP | BIND | 10822173 |
| DOK2 | p56DOK | p56dok-2 | docking protein 2, 56kDa | Affinity Capture-Western | BioGRID | 10822173 |
| DOK2 | p56DOK | p56dok-2 | docking protein 2, 56kDa | Dok2 interacts with RASGAP | BIND | 10822173 |
| DOK2 | p56DOK | p56dok-2 | docking protein 2, 56kDa | - | HPRD | 10508618 |10799545 |
| DOK4 | FLJ10488 | docking protein 4 | - | HPRD | 12730241 |
| DOK4 | FLJ10488 | docking protein 4 | IRS5/DOK4 interacts with RasGAP. This interaction was modelled on a demonstrated interaction between human IRS5/DOK4 and hamster RasGAP. | BIND | 12730241 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | Reconstituted Complex | BioGRID | 8647858 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | GAP interacts with EGFR. | BIND | 1385407 |1850098 |2176151 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | - | HPRD | 1633149 |
| EPHB2 | CAPB | DRT | EPHT3 | ERK | Hek5 | MGC87492 | PCBC | Tyro5 | EPH receptor B2 | - | HPRD,BioGRID | 9233798 |10644995 |
| EPHB3 | ETK2 | HEK2 | TYRO6 | EPH receptor B3 | - | HPRD,BioGRID | 9674711 |
| FES | FPS | feline sarcoma oncogene | - | HPRD,BioGRID | 7691175 |
| G3BP1 | G3BP | HDH-VIII | MGC111040 | GTPase activating protein (SH3 domain) binding protein 1 | - | HPRD,BioGRID | 9632780 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | RACK1 interacts with p120GAP.This interaction was modeled on a demonstrated interaction between RACK1 from mouse and rat, and human p120GAP. | BIND | 11350068 |
| GNB2L1 | Gnb2-rs1 | H12.3 | HLC-7 | PIG21 | RACK1 | guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | - | HPRD,BioGRID | 11350068 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD,BioGRID | 12441060 |
| GRLF1 | GRF-1 | KIAA1722 | MGC10745 | P190-A | P190A | p190RhoGAP | glucocorticoid receptor DNA binding factor 1 | Reconstituted Complex | BioGRID | 9034330 |
| HCK | JTK9 | hemopoietic cell kinase | - | HPRD,BioGRID | 7782336 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | - | HPRD,BioGRID | 7628625 |9219684 |
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | - | HPRD | 2833817 |7628625 |8663024 |9219684 |
| HSPD1 | CPN60 | GROEL | HSP60 | HSP65 | HuCHA60 | SPG13 | heat shock 60kDa protein 1 (chaperonin) | - | HPRD | 1347942 |
| HTT | HD | IT15 | huntingtin | HDP interacts with RasGAP. | BIND | 9079622 |
| HTT | HD | IT15 | huntingtin | - | HPRD,BioGRID | 9079622 |
| IGF1R | CD221 | IGFIR | JTK13 | MGC142170 | MGC142172 | MGC18216 | insulin-like growth factor 1 receptor | - | HPRD,BioGRID | 7642582 |
| INSR | CD220 | HHF5 | insulin receptor | - | HPRD,BioGRID | 1331107 |
| KDR | CD309 | FLK1 | VEGFR | VEGFR2 | kinase insert domain receptor (a type III receptor tyrosine kinase) | Affinity Capture-Western | BioGRID | 10319320 |
| KHDRBS1 | FLJ34027 | Sam68 | p62 | KH domain containing, RNA binding, signal transduction associated 1 | - | HPRD,BioGRID | 11604231 |
| KHDRBS1 | FLJ34027 | Sam68 | p62 | KH domain containing, RNA binding, signal transduction associated 1 | Sam68 interacts with the SH2 domains of p120GAP, preferentially with the C-terminal SH2 domain. This interaction was modelled based on a demonstrated interaction between rat proteins. | BIND | 11604231 |
| MAGI1 | AIP3 | BAIAP1 | BAP1 | MAGI-1 | TNRC19 | WWP3 | membrane associated guanylate kinase, WW and PDZ domain containing 1 | - | HPRD | 9169421 |
| MAP4K4 | FLH21957 | FLJ10410 | FLJ20373 | FLJ90111 | HGK | KIAA0687 | NIK | mitogen-activated protein kinase kinase kinase kinase 4 | - | HPRD,BioGRID | 10669731 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | - | HPRD | 10508618 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | Affinity Capture-Western | BioGRID | 9233798 |
| PAG1 | CBP | FLJ37858 | MGC138364 | PAG | phosphoprotein associated with glycosphingolipid microdomains 1 | - | HPRD | 10790433 |
| PDE6D | PDED | phosphodiesterase 6D, cGMP-specific, rod, delta | - | HPRD | 11786539 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | Activated PDGFR interacts with GAP. This interaction was modeled on a demonstrated interaction between human PDGFR and canine GAP. | BIND | 2157284 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | N-terminal SH2 domain of GAP interacts with PDGFR. This interaction was modeled on a demonstrated interaction between GAP from human and PDGFR from an unspecified species. | BIND | 1385407 |2173144 |7689724 |8344248 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | - | HPRD,BioGRID | 10697503 |11896619 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | Autophosphorylated PDGFR interacts with GAP. This interaction was modelled on a demonstrated interaction between human PDGFR and GAP from an unspecified species. | BIND | 8890167 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | - | HPRD | 1336372 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | Affinity Capture-Western | BioGRID | 9743338 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | - | HPRD,BioGRID | 8798684 |10708762 |10713673 |
| PTPRC | B220 | CD45 | CD45R | GP180 | LCA | LY5 | T200 | protein tyrosine phosphatase, receptor type, C | - | HPRD | 8570203 |
| PXN | FLJ16691 | paxillin | - | HPRD,BioGRID | 10092539 |
| RAB5A | RAB5 | RAB5A, member RAS oncogene family | - | HPRD | 11536198 |
| RAP1A | KREV-1 | KREV1 | RAP1 | SMGP21 | RAP1A, member of RAS oncogene family | in vivo | BioGRID | 2164710 |
| SLC9A2 | NHE2 | solute carrier family 9 (sodium/hydrogen exchanger), member 2 | - | HPRD,BioGRID | 10187839 |
| SOCS3 | ATOD4 | CIS3 | Cish3 | MGC71791 | SOCS-3 | SSI-3 | SSI3 | suppressor of cytokine signaling 3 | - | HPRD,BioGRID | 11331873 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD,BioGRID | 1717825 |
| SYN1 | SYN1a | SYN1b | SYNI | synapsin I | - | HPRD,BioGRID | 10899172 |
| WWP1 | AIP5 | DKFZp434D2111 | Tiul1 | hSDRP1 | WW domain containing E3 ubiquitin protein ligase 1 | - | HPRD | 9169421 |
| WWP2 | AIP2 | WWp2-like | WW domain containing E3 ubiquitin protein ligase 2 | - | HPRD | 9169421 |
| YES1 | HsT441 | P61-YES | Yes | c-yes | v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 | - | HPRD,BioGRID | 1544885 |
| ZAP70 | FLJ17670 | FLJ17679 | SRK | STD | TZK | ZAP-70 | zeta-chain (TCR) associated protein kinase 70kDa | - | HPRD,BioGRID | 7760813 |
Section V. Pathway annotation
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-1/206 | 105 | 112 | 1A,m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-132/212 | 473 | 479 | m8 | hsa-miR-212SZ | UAACAGUCUCCAGUCACGGCC |
| hsa-miR-132brain | UAACAGUCUACAGCCAUGGUCG | ||||
| miR-145 | 415 | 421 | m8 | hsa-miR-145 | GUCCAGUUUUCCCAGGAAUCCCUU |
| miR-153 | 125 | 132 | 1A,m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-182 | 334 | 341 | 1A,m8 | hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA |
| hsa-miR-182 | UUUGGCAAUGGUAGAACUCACA | ||||
| miR-21 | 179 | 185 | 1A | hsa-miR-21brain | UAGCUUAUCAGACUGAUGUUGA |
| hsa-miR-590 | GAGCUUAUUCAUAAAAGUGCAG | ||||
| miR-223 | 377 | 384 | 1A,m8 | hsa-miR-223 | UGUCAGUUUGUCAAAUACCCC |
| miR-24 | 238 | 244 | m8 | hsa-miR-24SZ | UGGCUCAGUUCAGCAGGAACAG |
| miR-30-5p | 700 | 707 | 1A,m8 | hsa-miR-30a-5p | UGUAAACAUCCUCGACUGGAAG |
| hsa-miR-30cbrain | UGUAAACAUCCUACACUCUCAGC | ||||
| hsa-miR-30dSZ | UGUAAACAUCCCCGACUGGAAG | ||||
| hsa-miR-30bSZ | UGUAAACAUCCUACACUCAGCU | ||||
| hsa-miR-30e-5p | UGUAAACAUCCUUGACUGGA | ||||
| miR-31 | 78 | 85 | 1A,m8 | hsa-miR-31 | AGGCAAGAUGCUGGCAUAGCUG |
| miR-320 | 620 | 627 | 1A,m8 | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-335 | 76 | 82 | m8 | hsa-miR-335brain | UCAAGAGCAAUAACGAAAAAUGU |
| miR-342 | 118 | 124 | m8 | hsa-miR-342brain | UCUCACACAGAAAUCGCACCCGUC |
| miR-374 | 517 | 523 | 1A | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-376 | 367 | 373 | 1A | hsa-miR-376a | AUCAUAGAGGAAAAUCCACGU |
| hsa-miR-376b | AUCAUAGAGGAAAAUCCAUGUU | ||||
| miR-377 | 117 | 123 | m8 | hsa-miR-377 | AUCACACAAAGGCAACUUUUGU |
| miR-448 | 126 | 132 | 1A | hsa-miR-448 | UUGCAUAUGUAGGAUGUCCCAU |
| miR-505 | 583 | 589 | 1A | hsa-miR-505 | GUCAACACUUGCUGGUUUCCUC |
| miR-96 | 335 | 341 | 1A | hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC |
| hsa-miR-96brain | UUUGGCACUAGCACAUUUUUGC | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.