Gene Page: SHH
Summary ?
| GeneID | 6469 |
| Symbol | SHH |
| Synonyms | HHG1|HLP3|HPE3|MCOPCB5|SMMCI|TPT|TPTPS |
| Description | sonic hedgehog |
| Reference | MIM:600725|HGNC:HGNC:10848|HPRD:02838| |
| Gene type | protein-coding |
| Map location | 7q36 |
| Pascal p-value | 0.502 |
| Fetal beta | -0.021 |
| DMG | 3 (# studies) |
| eGene | Putamen basal ganglia Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 8 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg11742909 | 7 | 155601876 | SHH | 5.047E-4 | -0.324 | 0.047 | DMG:Wockner_2014 |
| cg00577464 | 7 | 155604780 | SHH | -0.026 | 0.93 | DMG:Nishioka_2013 | |
| cg05820448 | 7 | 155605094 | SHH | 1.24E-8 | -0.018 | 5.03E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12719664 | 7 | 154998349 | SHH | ENSG00000164690.3 | 1.61749E-6 | 0.04 | 606618 | gtex_brain_putamen_basal |
| rs4960580 | 7 | 154998951 | SHH | ENSG00000164690.3 | 1.27595E-6 | 0.04 | 606016 | gtex_brain_putamen_basal |
| rs4960581 | 7 | 154998952 | SHH | ENSG00000164690.3 | 1.27415E-6 | 0.04 | 606015 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
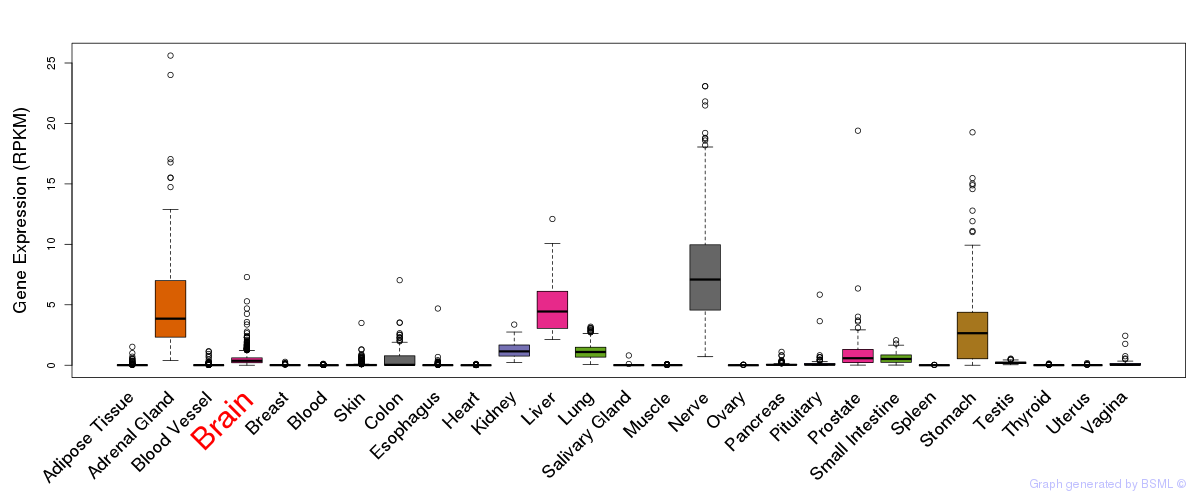
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA1468 | 0.87 | 0.89 |
| INPP5F | 0.87 | 0.89 |
| PCYOX1 | 0.86 | 0.90 |
| KIAA0317 | 0.86 | 0.90 |
| MAPRE2 | 0.86 | 0.90 |
| UBE2Z | 0.86 | 0.89 |
| UGCG | 0.85 | 0.88 |
| RTN4 | 0.85 | 0.88 |
| RABGEF1 | 0.85 | 0.87 |
| CLASP2 | 0.85 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.69 | -0.71 |
| MT-CO2 | -0.68 | -0.71 |
| FXYD1 | -0.68 | -0.68 |
| AF347015.21 | -0.67 | -0.71 |
| AF347015.8 | -0.67 | -0.70 |
| AF347015.33 | -0.67 | -0.67 |
| AF347015.2 | -0.66 | -0.68 |
| MT-CYB | -0.65 | -0.67 |
| AC021016.1 | -0.65 | -0.69 |
| AF347015.27 | -0.65 | -0.67 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005113 | patched binding | IEA | - | |
| GO:0005515 | protein binding | ISS | - | |
| GO:0008233 | peptidase activity | IEA | - | |
| GO:0043237 | laminin-1 binding | ISS | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007411 | axon guidance | ISS | axon (GO term level: 13) | - |
| GO:0048663 | neuron fate commitment | ISS | neuron (GO term level: 9) | - |
| GO:0021978 | telencephalon regionalization | IEA | Brain (GO term level: 10) | - |
| GO:0060020 | Bergmann glial cell differentiation | IEA | astrocyte, Glial (GO term level: 6) | - |
| GO:0030902 | hindbrain development | ISS | Brain (GO term level: 8) | - |
| GO:0030901 | midbrain development | ISS | Brain (GO term level: 8) | - |
| GO:0030900 | forebrain development | ISS | Brain (GO term level: 8) | - |
| GO:0014003 | oligodendrocyte development | IEA | axon, oligodendrocyte, Glial (GO term level: 10) | - |
| GO:0001755 | neural crest cell migration | ISS | - | |
| GO:0001570 | vasculogenesis | ISS | - | |
| GO:0001569 | patterning of blood vessels | ISS | - | |
| GO:0001658 | ureteric bud branching | ISS | - | |
| GO:0001656 | metanephros development | ISS | - | |
| GO:0001708 | cell fate specification | ISS | - | |
| GO:0002052 | positive regulation of neuroblast proliferation | IEA | - | |
| GO:0002076 | osteoblast development | IEA | - | |
| GO:0007267 | cell-cell signaling | ISS | - | |
| GO:0007228 | positive regulation of hh target transcription factor activity | IEA | - | |
| GO:0009952 | anterior/posterior pattern formation | IEA | - | |
| GO:0009887 | organ morphogenesis | ISS | - | |
| GO:0007165 | signal transduction | ISS | - | |
| GO:0007507 | heart development | ISS | - | |
| GO:0007442 | hindgut morphogenesis | IEA | - | |
| GO:0007435 | salivary gland morphogenesis | ISS | - | |
| GO:0007418 | ventral midline development | TAS | 8896572 | |
| GO:0007389 | pattern specification process | ISS | - | |
| GO:0008209 | androgen metabolic process | ISS | - | |
| GO:0030850 | prostate gland development | ISS | - | |
| GO:0031069 | hair follicle morphogenesis | IEA | - | |
| GO:0016539 | intein-mediated protein splicing | IEA | - | |
| GO:0042475 | odontogenesis of dentine-containing tooth | IEA | - | |
| GO:0051146 | striated muscle cell differentiation | IEA | - | |
| GO:0021940 | positive regulation of granule cell precursor proliferation | IEA | - | |
| GO:0021938 | smoothened signaling pathway involved in regulation of granule cell precursor cell proliferation | IEA | - | |
| GO:0042733 | embryonic digit morphogenesis | ISS | - | |
| GO:0042130 | negative regulation of T cell proliferation | IEA | - | |
| GO:0030539 | male genitalia development | ISS | - | |
| GO:0043010 | camera-type eye development | IEA | - | |
| GO:0032435 | negative regulation of proteasomal ubiquitin-dependent protein catabolic process | IEA | - | |
| GO:0030162 | regulation of proteolysis | ISS | - | |
| GO:0030336 | negative regulation of cell migration | ISS | - | |
| GO:0030324 | lung development | ISS | - | |
| GO:0021513 | spinal cord dorsal/ventral patterning | IEA | - | |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | IEA | - | |
| GO:0045596 | negative regulation of cell differentiation | ISS | - | |
| GO:0046639 | negative regulation of alpha-beta T cell differentiation | IEA | - | |
| GO:0048568 | embryonic organ development | IEA | - | |
| GO:0048589 | developmental growth | IEA | - | |
| GO:0048598 | embryonic morphogenesis | IEA | - | |
| GO:0045445 | myoblast differentiation | IEA | - | |
| GO:0045449 | regulation of transcription | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | ISS | - | |
| GO:0009986 | cell surface | ISS | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0045121 | membrane raft | ISS | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HEDGEHOG SIGNALING PATHWAY | 56 | 42 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG BASAL CELL CARCINOMA | 55 | 44 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SHH PATHWAY | 16 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PTC1 PATHWAY | 11 | 6 | All SZGR 2.0 genes in this pathway |
| PID HEDGEHOG 2PATHWAY | 22 | 17 | All SZGR 2.0 genes in this pathway |
| PID HEDGEHOG GLI PATHWAY | 48 | 35 | All SZGR 2.0 genes in this pathway |
| PID TAP63 PATHWAY | 54 | 40 | All SZGR 2.0 genes in this pathway |
| PID HNF3A PATHWAY | 44 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS B 2 SECRETIN FAMILY RECEPTORS | 88 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR LIGAND BINDING | 408 | 246 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN | 391 | 222 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 12HR UP | 116 | 79 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| WEBER METHYLATED IN COLON CANCER | 18 | 13 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR DN | 178 | 121 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K4ME3 AND H3K27ME3 | 210 | 148 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |