Gene Page: MRPS6
Summary ?
| GeneID | 64968 |
| Symbol | MRPS6 |
| Synonyms | C21orf101|MRP-S6|RPMS6|S6mt |
| Description | mitochondrial ribosomal protein S6 |
| Reference | MIM:611973|HGNC:HGNC:14051|Ensembl:ENSG00000243927|HPRD:11374|Vega:OTTHUMG00000065820 |
| Gene type | protein-coding |
| Map location | 21q22.11 |
| Pascal p-value | 0.01 |
| Sherlock p-value | 0.412 |
| Fetal beta | 0.602 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg06728306 | 21 | 35445589 | MRPS6;SLC5A3 | 5.58E-5 | 0.355 | 0.022 | DMG:Wockner_2014 |
| cg10599571 | 21 | 35445161 | MRPS6;SLC5A3 | 4.198E-4 | 0.344 | 0.045 | DMG:Wockner_2014 |
| cg04096688 | 21 | 35574180 | MRPS6 | 6.21E-9 | -0.018 | 3.28E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17049408 | chr2 | 58431128 | MRPS6 | 64968 | 0.19 | trans | ||
| rs11850361 | chr14 | 24220154 | MRPS6 | 64968 | 0.16 | trans | ||
| rs17793479 | chr20 | 59328916 | MRPS6 | 64968 | 0.2 | trans | ||
| rs2300390 | 21 | 35969515 | MRPS6 | ENSG00000243927.1 | 4.72385E-7 | 0.02 | 523991 | gtex_brain_ba24 |
| rs12482676 | 21 | 35970712 | MRPS6 | ENSG00000243927.1 | 8.6745E-7 | 0.02 | 525188 | gtex_brain_ba24 |
| rs915540 | 21 | 35985895 | MRPS6 | ENSG00000243927.1 | 7.79706E-7 | 0.02 | 540371 | gtex_brain_ba24 |
| rs12483565 | 21 | 35989970 | MRPS6 | ENSG00000243927.1 | 5.81351E-7 | 0.02 | 544446 | gtex_brain_ba24 |
Section II. Transcriptome annotation
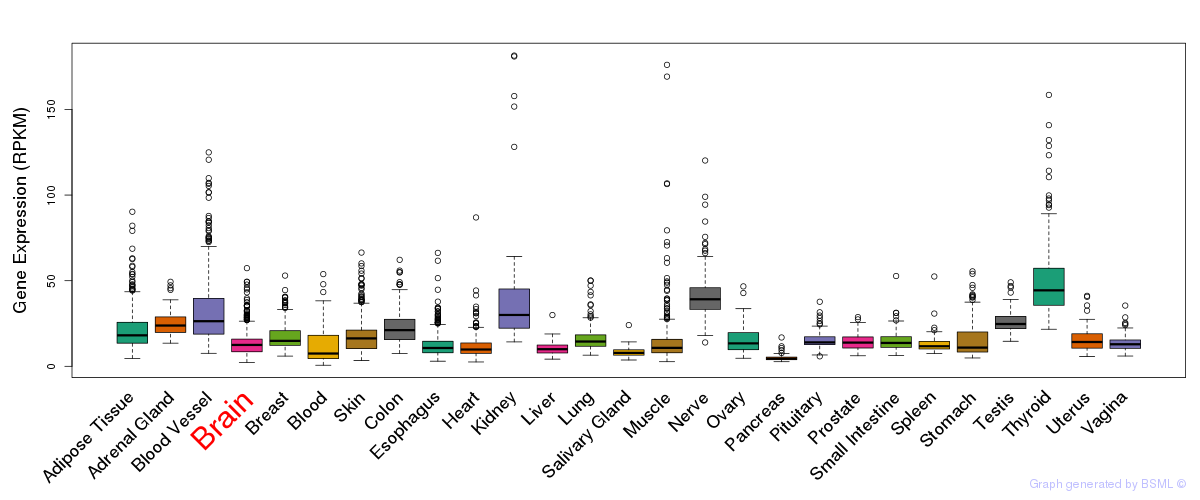
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| OSMAN BLADDER CANCER DN | 406 | 230 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| JIANG VHL TARGETS | 138 | 91 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE UP | 126 | 92 | All SZGR 2.0 genes in this pathway |
| HONMA DOCETAXEL RESISTANCE | 34 | 23 | All SZGR 2.0 genes in this pathway |
| NAKAMURA METASTASIS | 47 | 28 | All SZGR 2.0 genes in this pathway |
| MALONEY RESPONSE TO 17AAG UP | 41 | 26 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS EARLY UP | 66 | 44 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS LATE UP | 104 | 67 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA VIA VHL | 34 | 24 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS SENESCENT | 572 | 352 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| GHANDHI DIRECT IRRADIATION DN | 33 | 23 | All SZGR 2.0 genes in this pathway |