Gene Page: SNRPD3
Summary ?
| GeneID | 6634 |
| Symbol | SNRPD3 |
| Synonyms | SMD3|Sm-D3 |
| Description | small nuclear ribonucleoprotein D3 polypeptide |
| Reference | MIM:601062|HGNC:HGNC:11160|HPRD:03039| |
| Gene type | protein-coding |
| Map location | 22q11.23 |
| Pascal p-value | 0.108 |
| Sherlock p-value | 0.974 |
| Fetal beta | 0.671 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg21703572 | 22 | 24951237 | SNRPD3 | -0.024 | 0.39 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10182691 | chr2 | 34122007 | SNRPD3 | 6634 | 0.05 | trans | ||
| rs7595584 | chr2 | 175652187 | SNRPD3 | 6634 | 0.1 | trans | ||
| rs17146240 | chr5 | 119522933 | SNRPD3 | 6634 | 0 | trans | ||
| rs17146261 | chr5 | 119534252 | SNRPD3 | 6634 | 0 | trans | ||
| rs6032295 | chr20 | 44178138 | SNRPD3 | 6634 | 0.04 | trans | ||
| rs5952167 | chrX | 115529371 | SNRPD3 | 6634 | 0.07 | trans |
Section II. Transcriptome annotation
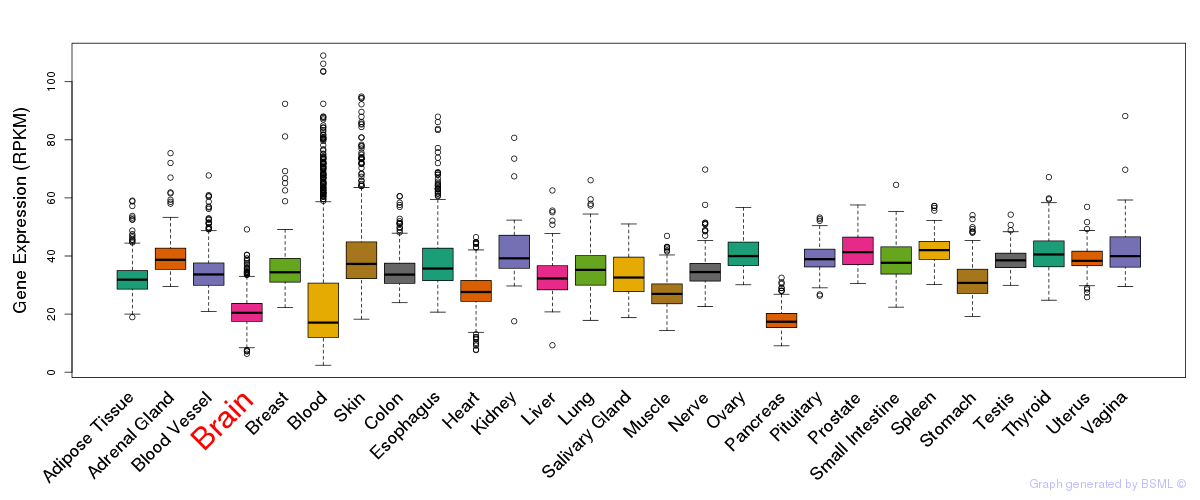
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DNAH7 | 0.82 | 0.73 |
| LRRC6 | 0.80 | 0.74 |
| TSGA10 | 0.79 | 0.58 |
| WDR66 | 0.79 | 0.67 |
| UBXN10 | 0.78 | 0.69 |
| CASC1 | 0.77 | 0.61 |
| AC079354.2 | 0.77 | 0.56 |
| CCDC39 | 0.76 | 0.65 |
| AK7 | 0.76 | 0.62 |
| NPHP1 | 0.75 | 0.58 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.34 | -0.17 |
| AF347015.2 | -0.31 | -0.17 |
| MT-CO2 | -0.30 | -0.18 |
| AF347015.18 | -0.30 | -0.13 |
| AF347015.8 | -0.29 | -0.15 |
| AC098691.2 | -0.29 | -0.25 |
| AF347015.31 | -0.28 | -0.18 |
| MT-CYB | -0.27 | -0.14 |
| AF347015.15 | -0.26 | -0.13 |
| AF347015.33 | -0.26 | -0.13 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 11714716 |11748230 |12065586 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000387 | spliceosomal snRNP biogenesis | EXP | 15130578 | |
| GO:0008380 | RNA splicing | TAS | 1701240 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 10531003 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005654 | nucleoplasm | EXP | 10531003 | |
| GO:0005681 | spliceosome | TAS | 7527560 | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0030532 | small nuclear ribonucleoprotein complex | TAS | 1701240 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CLNS1A | CLCI | CLNS1B | ICln | chloride channel, nucleotide-sensitive, 1A | Affinity Capture-Western Reconstituted Complex | BioGRID | 11713266 |
| DDX20 | DKFZp434H052 | DP103 | GEMIN3 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 20 | - | HPRD,BioGRID | 10601333 |
| GEMIN5 | DKFZp586M1824 | MGC142174 | gem (nuclear organelle) associated protein 5 | - | HPRD,BioGRID | 11714716 |
| GEMIN6 | FLJ23459 | gem (nuclear organelle) associated protein 6 | Gemin6 interacts with SNRPD3 (SmD3) | BIND | 15939020 |
| GEMIN6 | FLJ23459 | gem (nuclear organelle) associated protein 6 | Gemin6 interacts with SmD3. | BIND | 11748230 |
| GEMIN7 | SIP3 | gem (nuclear organelle) associated protein 7 | GEMIN7 interacts with SNRPD3 (SmD3) | BIND | 12065586 |
| LSM10 | MGC15749 | MST074 | MSTP074 | LSM10, U7 small nuclear RNA associated | Affinity Capture-MS | BioGRID | 12975319 |
| LSM3 | SMX4 | USS2 | YLR438C | LSM3 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD,BioGRID | 15231747 |
| LSM7 | YNL147W | LSM7 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD,BioGRID | 15231747 |
| PRMT5 | HRMT1L5 | IBP72 | JBP1 | SKB1 | SKB1Hs | protein arginine methyltransferase 5 | - | HPRD,BioGRID | 11713266 |
| RNU11 | U11 | RNA, U11 small nuclear | Affinity Capture-MS | BioGRID | 15146077 |
| RNU12 | U12 | RNA, U12 small nuclear | Affinity Capture-MS | BioGRID | 15146077 |
| SF3A2 | PRP11 | PRPF11 | SAP62 | SF3a66 | splicing factor 3a, subunit 2, 66kDa | Affinity Capture-MS | BioGRID | 12234937 |
| SFRS12 | DKFZp564B176 | MGC133045 | SRrp508 | SRrp86 | splicing factor, arginine/serine-rich 12 | - | HPRD | 14559993 |
| SMN1 | BCD541 | SMA | SMA1 | SMA2 | SMA3 | SMA4 | SMA@ | SMN | SMNT | T-BCD541 | survival of motor neuron 1, telomeric | Reconstituted Complex | BioGRID | 10851237 |
| SMN2 | BCD541 | C-BCD541 | FLJ76644 | MGC20996 | MGC5208 | SMNC | survival of motor neuron 2, centromeric | - | HPRD | 9323129 |
| SNRPB | COD | SNRPB1 | SmB/SmB' | snRNP-B | small nuclear ribonucleoprotein polypeptides B and B1 | - | HPRD,BioGRID | 10025403 |
| SNRPE | B-raf | SME | small nuclear ribonucleoprotein polypeptide E | Two-hybrid | BioGRID | 9417867 |
| SNRPE | B-raf | SME | small nuclear ribonucleoprotein polypeptide E | SNRPD3 interacts with SNRPE. | BIND | 9417867 |
| SNRPG | MGC117317 | SMG | small nuclear ribonucleoprotein polypeptide G | Two-hybrid | BioGRID | 9417867 |
| SNRPG | MGC117317 | SMG | small nuclear ribonucleoprotein polypeptide G | SNRPD3 interacts with SNRPG. | BIND | 9417867 |
| SNRPN | DKFZp686C0927 | DKFZp686M12165 | DKFZp761I1912 | DKFZp762N022 | FLJ33569 | FLJ36996 | FLJ39265 | HCERN3 | MGC29886 | PWCR | RT-LI | SM-D | SMN | SNRNP-N | SNURF-SNRPN | small nuclear ribonucleoprotein polypeptide N | SNRPD3 interacts with SNRPN. | BIND | 9417867 |
| SNRPN | DKFZp686C0927 | DKFZp686M12165 | DKFZp761I1912 | DKFZp762N022 | FLJ33569 | FLJ36996 | FLJ39265 | HCERN3 | MGC29886 | PWCR | RT-LI | SM-D | SMN | SNRNP-N | SNURF-SNRPN | small nuclear ribonucleoprotein polypeptide N | Two-hybrid | BioGRID | 9417867 |
| STXBP3 | MUNC18-3 | MUNC18C | PSP | UNC-18C | syntaxin binding protein 3 | - | HPRD | 12065586 |
| WDR77 | HKMT1069 | MEP50 | MGC2722 | Nbla10071 | RP11-552M11.3 | p44 | WD repeat domain 77 | - | HPRD,BioGRID | 11756452 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SPLICEOSOME | 128 | 72 | All SZGR 2.0 genes in this pathway |
| KEGG SYSTEMIC LUPUS ERYTHEMATOSUS | 140 | 100 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF NON CODING RNA | 49 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | 140 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME SLBP DEPENDENT PROCESSING OF REPLICATION DEPENDENT HISTONE PRE MRNAS | 11 | 5 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION | 105 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA PROCESSING | 161 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA SPLICING | 111 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA SPLICING MINOR PATHWAY | 45 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME PROCESSING OF CAPPED INTRONLESS PRE MRNA | 23 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF RNA | 330 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | 44 | 22 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| OUELLET OVARIAN CANCER INVASIVE VS LMP UP | 117 | 85 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| MATTIOLI MGUS VS PCL | 116 | 62 | All SZGR 2.0 genes in this pathway |
| JAERVINEN AMPLIFIED IN LARYNGEAL CANCER | 40 | 24 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| FERREIRA EWINGS SARCOMA UNSTABLE VS STABLE UP | 167 | 92 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 12HR UP | 116 | 79 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA SUBGROUPS | 30 | 20 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C4 | 20 | 14 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE DN | 123 | 76 | All SZGR 2.0 genes in this pathway |
| CLAUS PGR POSITIVE MENINGIOMA UP | 10 | 7 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL OPTIMAL DEBULKING | 246 | 152 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| CHAUHAN RESPONSE TO METHOXYESTRADIOL UP | 51 | 32 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC UP | 72 | 53 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS UP | 143 | 100 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 14 | 143 | 86 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM SENSITIVE VIA TSC2 | 39 | 25 | All SZGR 2.0 genes in this pathway |
| KIM TIAL1 TARGETS | 32 | 22 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| ABRAMSON INTERACT WITH AIRE | 45 | 33 | All SZGR 2.0 genes in this pathway |