Gene Page: BTK
Summary ?
| GeneID | 695 |
| Symbol | BTK |
| Synonyms | AGMX1|AT|ATK|BPK|IMD1|PSCTK1|XLA |
| Description | Bruton tyrosine kinase |
| Reference | MIM:300300|HGNC:HGNC:1133|Ensembl:ENSG00000010671|HPRD:02248|Vega:OTTHUMG00000022022 |
| Gene type | protein-coding |
| Map location | Xq21.33-q22 |
| Fetal beta | -0.881 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Kranz_2015 | Whole Exome Sequencing and Targeted Sequencing | DNMs in 5 genes were identified through WES and independently validated through TS in this study | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.1022 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BTK | chrX | 100615568 | C | T | NM_000061 | p.R255Q | nonsynonymous SNV | 1.286 | P | Schizophrenia | DNM:Kranz_2015 |
Section II. Transcriptome annotation
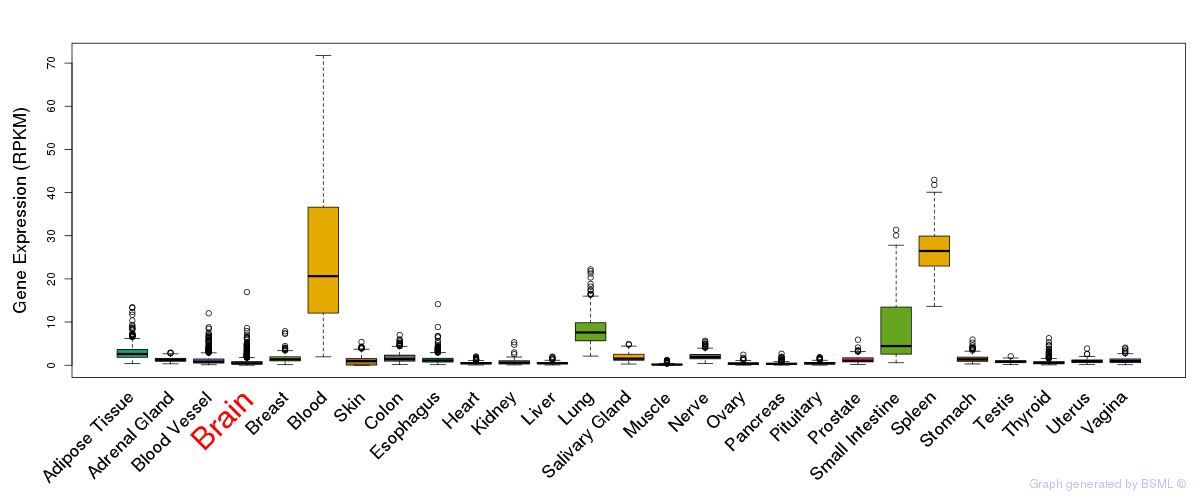
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NUSAP1 | 0.99 | 0.85 |
| TPX2 | 0.99 | 0.83 |
| KIF4A | 0.99 | 0.84 |
| BUB1B | 0.99 | 0.86 |
| CCNB2 | 0.99 | 0.87 |
| MELK | 0.99 | 0.84 |
| KIF23 | 0.99 | 0.80 |
| PRC1 | 0.98 | 0.66 |
| CCNA2 | 0.98 | 0.84 |
| EXO1 | 0.98 | 0.78 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SLC9A3R2 | -0.40 | -0.55 |
| FBXO2 | -0.40 | -0.71 |
| HLA-F | -0.39 | -0.77 |
| PTH1R | -0.39 | -0.71 |
| C5orf53 | -0.39 | -0.69 |
| ASPHD1 | -0.38 | -0.59 |
| TNFSF12 | -0.38 | -0.68 |
| ALDOC | -0.38 | -0.73 |
| LHPP | -0.38 | -0.63 |
| AIFM3 | -0.37 | -0.72 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0005524 | ATP binding | TAS | 11913944 | |
| GO:0004713 | protein tyrosine kinase activity | IEA | - | |
| GO:0004715 | non-membrane spanning protein tyrosine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0016301 | kinase activity | IEA | - | |
| GO:0042802 | identical protein binding | IPI | 11577348 | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007242 | intracellular signaling cascade | IEA | - | |
| GO:0007242 | intracellular signaling cascade | TAS | 11913944 | |
| GO:0007498 | mesoderm development | TAS | 8013627 | |
| GO:0007249 | I-kappaB kinase/NF-kappaB cascade | IEA | - | |
| GO:0008624 | induction of apoptosis by extracellular signals | TAS | 8688094 | |
| GO:0019722 | calcium-mediated signaling | TAS | 15046600 | |
| GO:0018108 | peptidyl-tyrosine phosphorylation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IDA | 18029348 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IDA | 18029348 | |
| GO:0005737 | cytoplasm | TAS | 15046600 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0031410 | cytoplasmic vesicle | IEA | - | |
| GO:0045121 | membrane raft | IDA | 15046600 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARHGAP17 | DKFZp564A1363 | FLJ37567 | FLJ43368 | MGC87805 | MST066 | MST110 | MSTP038 | MSTP066 | MSTP110 | NADRIN | RICH1 | WBP15 | Rho GTPase activating protein 17 | Reconstituted Complex | BioGRID | 11431473 |
| ARID3A | BRIGHT | DRIL1 | DRIL3 | E2FBP1 | AT rich interactive domain 3A (BRIGHT-like) | - | HPRD,BioGRID | 15203319 |
| BLNK | BASH | BLNK-S | LY57 | MGC111051 | SLP-65 | SLP65 | B-cell linker | - | HPRD | 10498607 |10556826 |
| BLNK | BASH | BLNK-S | LY57 | MGC111051 | SLP-65 | SLP65 | B-cell linker | Affinity Capture-Western in vitro in vivo | BioGRID | 10498607 |12093870 |
| BTK | AGMX1 | AT | ATK | BPK | IMD1 | MGC126261 | MGC126262 | PSCTK1 | XLA | Bruton agammaglobulinemia tyrosine kinase | - | HPRD,BioGRID | 11527964 |
| CAV1 | CAV | MSTP085 | VIP21 | caveolin 1, caveolae protein, 22kDa | - | HPRD,BioGRID | 11751885 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | - | HPRD,BioGRID | 7629518 |10427990 |
| CBLB | DKFZp686J10223 | DKFZp779A0729 | DKFZp779F1443 | FLJ36865 | FLJ41152 | Nbla00127 | RNF56 | Cas-Br-M (murine) ecotropic retroviral transforming sequence b | Affinity Capture-Western | BioGRID | 12093870 |
| EWSR1 | EWS | Ewing sarcoma breakpoint region 1 | - | HPRD,BioGRID | 9201297 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | - | HPRD | 9751072 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | Reconstituted Complex | BioGRID | 8058772 |
| GNA12 | MGC104623 | MGC99644 | NNX3 | RMP | gep | guanine nucleotide binding protein (G protein) alpha 12 | in vitro in vivo | BioGRID | 9796816 |
| GNAQ | G-ALPHA-q | GAQ | guanine nucleotide binding protein (G protein), q polypeptide | - | HPRD,BioGRID | 9770463 |
| GNG2 | - | guanine nucleotide binding protein (G protein), gamma 2 | - | HPRD | 7972043 |11698416 |
| GTF2I | BAP-135 | BAP135 | BTKAP1 | DIWS | FLJ38776 | FLJ56355 | IB291 | SPIN | TFII-I | WBS | WBSCR6 | general transcription factor II, i | - | HPRD,BioGRID | 9012831 |10373551 |14623887 |
| HCK | JTK9 | hemopoietic cell kinase | - | HPRD,BioGRID | 8058772 |
| IBTK | BTKI | KIAA1417 | MGC142256 | MGC142258 | RP1-93K22.1 | inhibitor of Bruton agammaglobulinemia tyrosine kinase | - | HPRD,BioGRID | 11577348 |
| JAK1 | JAK1A | JAK1B | JTK3 | Janus kinase 1 (a protein tyrosine kinase) | - | HPRD,BioGRID | 9178903 |
| KHDRBS1 | FLJ34027 | Sam68 | p62 | KH domain containing, RNA binding, signal transduction associated 1 | - | HPRD,BioGRID | 9201297 |
| LYN | FLJ26625 | JTK8 | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | - | HPRD,BioGRID | 8058772 |
| PLCG2 | - | phospholipase C, gamma 2 (phosphatidylinositol-specific) | Affinity Capture-Western | BioGRID | 10981967 |12093870 |
| PRKCB | MGC41878 | PKC-beta | PKCB | PRKCB1 | PRKCB2 | protein kinase C, beta | Reconstituted Complex | BioGRID | 12679936 |
| PRKD1 | PKC-MU | PKCM | PKD | PRKCM | protein kinase D1 | - | HPRD,BioGRID | 10561498 |
| RPS6KB1 | PS6K | S6K | S6K1 | STK14A | p70(S6K)-alpha | p70-S6K | p70-alpha | ribosomal protein S6 kinase, 70kDa, polypeptide 1 | Biochemical Activity | BioGRID | 16640565 |
| RPS6KB2 | KLS | P70-beta | P70-beta-1 | P70-beta-2 | S6K-beta2 | S6K2 | SRK | STK14B | p70(S6K)-beta | p70S6Kb | ribosomal protein S6 kinase, 70kDa, polypeptide 2 | Biochemical Activity | BioGRID | 16640565 |
| SH2B2 | APS | SH2B adaptor protein 2 | - | HPRD | 10872802 |
| SH3BP5 | SAB | SH3-domain binding protein 5 (BTK-associated) | - | HPRD,BioGRID | 9571151 |
| SH3BP5 | SAB | SH3-domain binding protein 5 (BTK-associated) | - | HPRD | 9571151 |10339589 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | - | HPRD,BioGRID | 9201297 |
| WAS | IMD2 | THC | WASP | Wiskott-Aldrich syndrome (eczema-thrombocytopenia) | - | HPRD,BioGRID | 8892607 |
| WASF2 | SCAR2 | WAVE2 | dJ393P12.2 | WAS protein family, member 2 | - | HPRD | 10427990 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG B CELL RECEPTOR SIGNALING PATHWAY | 75 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG FC EPSILON RI SIGNALING PATHWAY | 79 | 58 | All SZGR 2.0 genes in this pathway |
| KEGG PRIMARY IMMUNODEFICIENCY | 35 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BCR PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FCER1 PATHWAY | 41 | 30 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PTDINS PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| ST GA12 PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| SIG CHEMOTAXIS | 45 | 37 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN B LYMPHOCYTES | 36 | 31 | All SZGR 2.0 genes in this pathway |
| SIG BCR SIGNALING PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| ST B CELL ANTIGEN RECEPTOR | 40 | 32 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | 37 | 29 | All SZGR 2.0 genes in this pathway |
| PID FCER1 PATHWAY | 62 | 43 | All SZGR 2.0 genes in this pathway |
| PID BCR 5PATHWAY | 65 | 50 | All SZGR 2.0 genes in this pathway |
| PID FAS PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| PID PI3KCI PATHWAY | 49 | 40 | All SZGR 2.0 genes in this pathway |
| PID EPO PATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | 29 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | 126 | 90 | All SZGR 2.0 genes in this pathway |
| REACTOME MYD88 MAL CASCADE INITIATED ON PLASMA MEMBRANE | 83 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED TLR4 SIGNALLING | 93 | 69 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME TOLL RECEPTOR CASCADES | 118 | 84 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 UP | 201 | 125 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| KLEIN TARGETS OF BCR ABL1 FUSION | 45 | 34 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS DN | 124 | 79 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE DN | 245 | 154 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| LEE METASTASIS AND ALTERNATIVE SPLICING UP | 74 | 51 | All SZGR 2.0 genes in this pathway |
| FINETTI BREAST CANCER KINOME GREEN | 16 | 14 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS UP | 102 | 64 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| VILIMAS NOTCH1 TARGETS DN | 20 | 12 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER UP | 288 | 168 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |