Gene Page: TIMP4
Summary ?
| GeneID | 7079 |
| Symbol | TIMP4 |
| Synonyms | - |
| Description | TIMP metallopeptidase inhibitor 4 |
| Reference | MIM:601915|HGNC:HGNC:11823|Ensembl:ENSG00000157150|HPRD:03557|Vega:OTTHUMG00000129763 |
| Gene type | protein-coding |
| Map location | 3p25 |
| Pascal p-value | 0.013 |
| Fetal beta | -0.975 |
| eGene | Frontal Cortex BA9 Hypothalamus Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.006 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2502827 | chr1 | 176044216 | TIMP4 | 7079 | 0.12 | trans | ||
| rs17572651 | chr1 | 218943612 | TIMP4 | 7079 | 2.206E-6 | trans | ||
| snp_a-1919794 | 0 | TIMP4 | 7079 | 0.01 | trans | |||
| rs16829545 | chr2 | 151977407 | TIMP4 | 7079 | 1.015E-10 | trans | ||
| rs3845734 | chr2 | 171125572 | TIMP4 | 7079 | 0 | trans | ||
| rs8179648 | chr2 | 179377279 | TIMP4 | 7079 | 0.07 | trans | ||
| rs7584986 | chr2 | 184111432 | TIMP4 | 7079 | 1.756E-18 | trans | ||
| rs6729020 | chr2 | 189064719 | TIMP4 | 7079 | 0.07 | trans | ||
| rs6807632 | chr3 | 111395567 | TIMP4 | 7079 | 0.12 | trans | ||
| rs10491487 | chr5 | 80323367 | TIMP4 | 7079 | 2.532E-7 | trans | ||
| rs1560919 | chr5 | 89587250 | TIMP4 | 7079 | 0.18 | trans | ||
| rs1368303 | chr5 | 147672388 | TIMP4 | 7079 | 2.362E-9 | trans | ||
| rs1929769 | chr6 | 40765977 | TIMP4 | 7079 | 0 | trans | ||
| rs17071719 | chr6 | 110793869 | TIMP4 | 7079 | 0.14 | trans | ||
| rs6989594 | chr8 | 126303866 | TIMP4 | 7079 | 0.13 | trans | ||
| rs11139334 | chr9 | 84209393 | TIMP4 | 7079 | 0.01 | trans | ||
| rs12099529 | chr12 | 78888019 | TIMP4 | 7079 | 0.07 | trans | ||
| rs7996604 | chr13 | 42224412 | TIMP4 | 7079 | 0.14 | trans | ||
| snp_a-1880548 | 0 | TIMP4 | 7079 | 0.05 | trans | |||
| rs17104720 | chr14 | 77127308 | TIMP4 | 7079 | 0.04 | trans | ||
| rs6574467 | chr14 | 79179744 | TIMP4 | 7079 | 0 | trans | ||
| rs10146003 | chr14 | 79191170 | TIMP4 | 7079 | 0 | trans | ||
| rs16955618 | chr15 | 29937543 | TIMP4 | 7079 | 1.848E-28 | trans | ||
| snp_a-1830894 | 0 | TIMP4 | 7079 | 0 | trans | |||
| snp_a-2175755 | 0 | TIMP4 | 7079 | 0.02 | trans | |||
| rs16944856 | chr17 | 11394566 | TIMP4 | 7079 | 0.1 | trans | ||
| rs17085767 | chr18 | 69839397 | TIMP4 | 7079 | 3.644E-4 | trans | ||
| rs749043 | chr20 | 46995700 | TIMP4 | 7079 | 0.02 | trans |
Section II. Transcriptome annotation
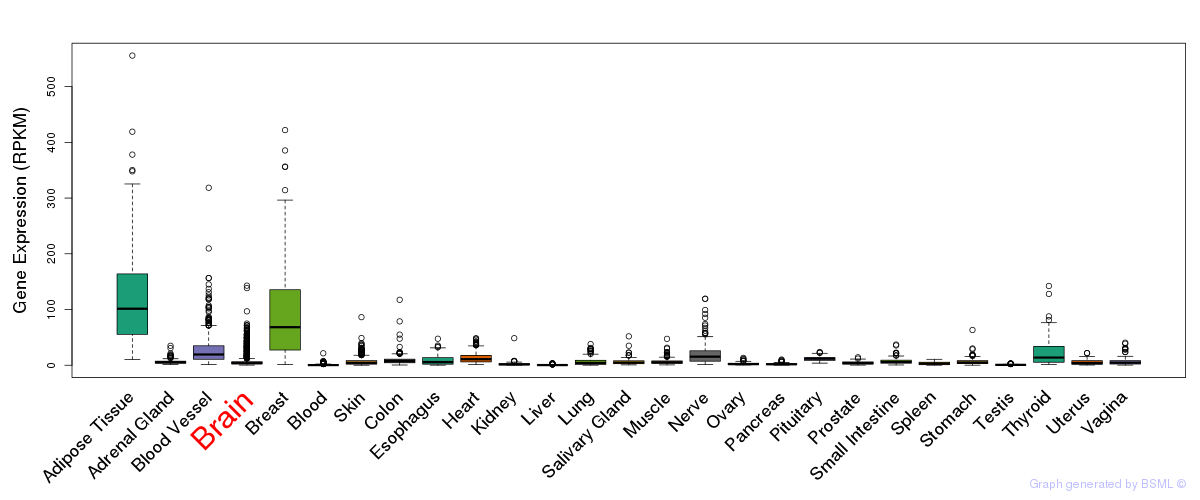
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ICAM2 | 0.84 | 0.69 |
| TMEM204 | 0.79 | 0.67 |
| GIMAP5 | 0.78 | 0.61 |
| C10orf10 | 0.78 | 0.49 |
| IL32 | 0.77 | 0.54 |
| SH3TC1 | 0.74 | 0.53 |
| CDC42EP5 | 0.70 | 0.72 |
| HRCT1 | 0.70 | 0.32 |
| GGT5 | 0.70 | 0.56 |
| RRAS | 0.68 | 0.59 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GGNBP2 | -0.56 | -0.57 |
| DNAJC7 | -0.55 | -0.53 |
| DNM1L | -0.54 | -0.53 |
| AC141586.1 | -0.53 | -0.56 |
| SLC4A1AP | -0.53 | -0.49 |
| TAOK3 | -0.53 | -0.51 |
| MPHOSPH8 | -0.53 | -0.57 |
| DDX42 | -0.53 | -0.51 |
| RP11-298P3.1 | -0.52 | -0.55 |
| SDCCAG1 | -0.52 | -0.56 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| GO:0004857 | enzyme inhibitor activity | IEA | - | |
| GO:0008191 | metalloendopeptidase inhibitor activity | NAS | 8939999 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008150 | biological_process | ND | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | NAS | - | |
| GO:0005578 | proteinaceous extracellular matrix | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 3 4WK DN | 39 | 27 | All SZGR 2.0 genes in this pathway |
| GOUYER TATI TARGETS DN | 17 | 12 | All SZGR 2.0 genes in this pathway |
| WOTTON RUNX TARGETS UP | 21 | 14 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| CHEN LVAD SUPPORT OF FAILING HEART UP | 103 | 69 | All SZGR 2.0 genes in this pathway |
| SAGIV CD24 TARGETS DN | 46 | 26 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE UP | 97 | 61 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN LUNG DN | 37 | 22 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL A UP | 84 | 52 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| BASSO HAIRY CELL LEUKEMIA DN | 80 | 66 | All SZGR 2.0 genes in this pathway |
| ZHENG GLIOBLASTOMA PLASTICITY UP | 250 | 168 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| BHATI G2M ARREST BY 2METHOXYESTRADIOL DN | 127 | 75 | All SZGR 2.0 genes in this pathway |
| RUIZ TNC TARGETS UP | 153 | 107 | All SZGR 2.0 genes in this pathway |
| LIU VAV3 PROSTATE CARCINOGENESIS UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS LCP WITH H3K4ME3 | 174 | 100 | All SZGR 2.0 genes in this pathway |
| NAKAYAMA SOFT TISSUE TUMORS PCA2 DN | 80 | 51 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES LCP WITH H3K4ME3 | 142 | 80 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC LCP WITH H3K4ME3 | 58 | 34 | All SZGR 2.0 genes in this pathway |
| NAKAMURA ADIPOGENESIS LATE UP | 104 | 67 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |
| NABA ECM REGULATORS | 238 | 125 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |