Gene Page: CAPN5
Summary ?
| GeneID | 726 |
| Symbol | CAPN5 |
| Synonyms | ADNIV|HTRA3|VRNI|nCL-3 |
| Description | calpain 5 |
| Reference | MIM:602537|HGNC:HGNC:1482|Ensembl:ENSG00000149260|HPRD:03963|Vega:OTTHUMG00000165192 |
| Gene type | protein-coding |
| Map location | 11q14 |
| Pascal p-value | 0.112 |
| Fetal beta | -0.282 |
| Support | RNA AND PROTEIN SYNTHESIS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CAPN5 | chr11 | 76831955 | G | A | NM_004055 NM_004055 | . p.496R>Q | splice missense | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
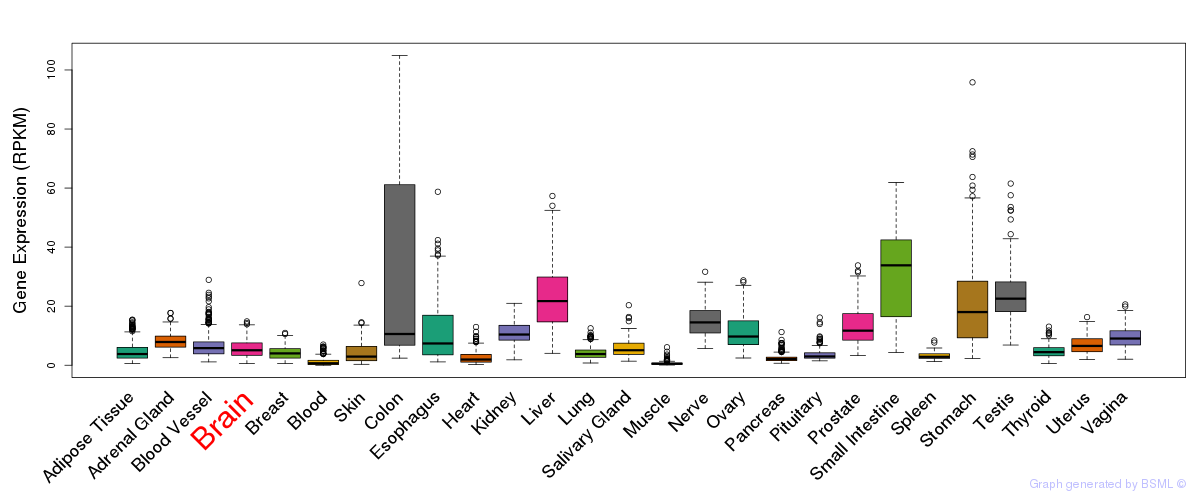
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LUM | 0.86 | 0.73 |
| ASPN | 0.86 | 0.81 |
| ITIH2 | 0.84 | 0.84 |
| ALDH1A2 | 0.82 | 0.83 |
| COL3A1 | 0.82 | 0.38 |
| OLFML3 | 0.82 | 0.58 |
| LYVE1 | 0.80 | 0.83 |
| TBX18 | 0.80 | 0.74 |
| OGN | 0.80 | 0.90 |
| BMP5 | 0.77 | 0.52 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ACP6 | -0.23 | -0.12 |
| SEPT12 | -0.22 | -0.33 |
| AL022328.1 | -0.22 | -0.19 |
| CHMP4A | -0.21 | -0.33 |
| SNHG12 | -0.21 | -0.39 |
| AC132872.1 | -0.20 | -0.42 |
| ZRSR2 | -0.19 | -0.34 |
| AMY2B | -0.18 | -0.12 |
| LRRC37A | -0.18 | -0.26 |
| AC007216.4 | -0.18 | -0.21 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BERTUCCI MEDULLARY VS DUCTAL BREAST CANCER DN | 169 | 118 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA DN | 349 | 157 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS UP | 238 | 144 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 11Q12 Q14 AMPLICON | 158 | 93 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| BRUNO HEMATOPOIESIS | 66 | 48 | All SZGR 2.0 genes in this pathway |
| LINDVALL IMMORTALIZED BY TERT DN | 80 | 56 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY DEACETYLATION IN PANCREATIC CANCER | 50 | 30 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| COATES MACROPHAGE M1 VS M2 UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| QI HYPOXIA | 140 | 96 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF | 367 | 231 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR TARGETS UP | 48 | 33 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |