Gene Page: TXK
Summary ?
| GeneID | 7294 |
| Symbol | TXK |
| Synonyms | BTKL|PSCTK5|PTK4|RLK|TKL |
| Description | TXK tyrosine kinase |
| Reference | MIM:600058|HGNC:HGNC:12434|Ensembl:ENSG00000074966|HPRD:02505|Vega:OTTHUMG00000102065 |
| Gene type | protein-coding |
| Map location | 4p12 |
| Pascal p-value | 0.067 |
| Fetal beta | 0.311 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
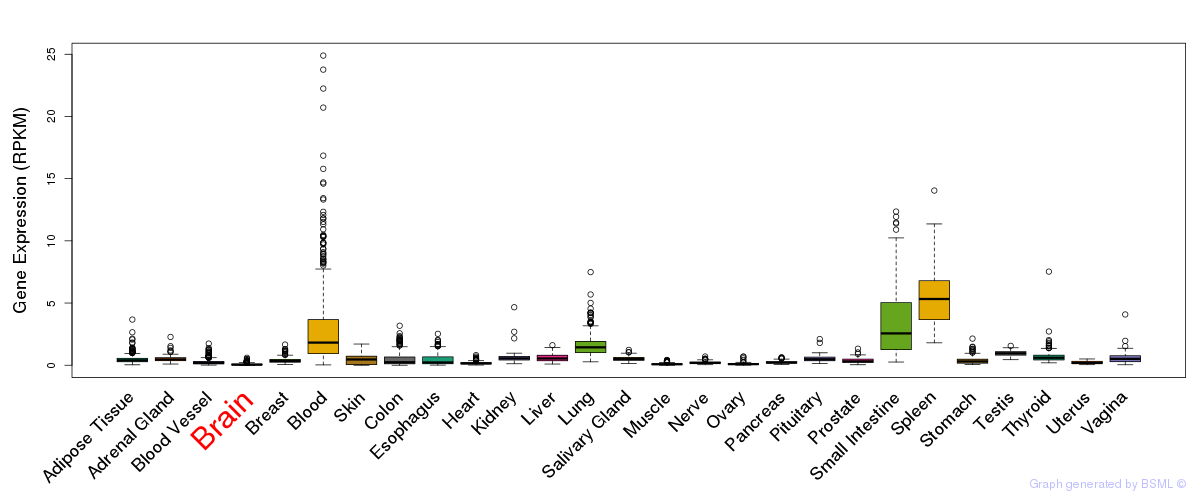
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CSNK1G3 | 0.97 | 0.97 |
| IPO7 | 0.96 | 0.97 |
| FYTTD1 | 0.96 | 0.97 |
| CAND1 | 0.96 | 0.97 |
| WDR47 | 0.95 | 0.96 |
| EXOC5 | 0.95 | 0.96 |
| SMEK2 | 0.95 | 0.96 |
| PPP2R5E | 0.95 | 0.97 |
| KPNA4 | 0.95 | 0.95 |
| BCLAF1 | 0.95 | 0.96 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FXYD1 | -0.76 | -0.85 |
| MT-CO2 | -0.75 | -0.84 |
| AF347015.31 | -0.75 | -0.83 |
| AF347015.33 | -0.74 | -0.82 |
| HIGD1B | -0.73 | -0.84 |
| IFI27 | -0.73 | -0.83 |
| MT-CYB | -0.72 | -0.81 |
| TSC22D4 | -0.72 | -0.79 |
| AF347015.8 | -0.71 | -0.82 |
| HSD17B14 | -0.71 | -0.77 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG LEUKOCYTE TRANSENDOTHELIAL MIGRATION | 118 | 78 | All SZGR 2.0 genes in this pathway |
| TAKEDA TARGETS OF NUP98 HOXA9 FUSION 3D UP | 182 | 110 | All SZGR 2.0 genes in this pathway |
| HAHTOLA SEZARY SYNDROM DN | 42 | 26 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE UP | 86 | 57 | All SZGR 2.0 genes in this pathway |
| SHIPP DLBCL VS FOLLICULAR LYMPHOMA DN | 45 | 28 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT DN | 129 | 86 | All SZGR 2.0 genes in this pathway |
| JOSEPH RESPONSE TO SODIUM BUTYRATE DN | 64 | 45 | All SZGR 2.0 genes in this pathway |
| VISALA AGING LYMPHOCYTE DN | 19 | 10 | All SZGR 2.0 genes in this pathway |
| FIRESTEIN PROLIFERATION | 175 | 125 | All SZGR 2.0 genes in this pathway |
| MARSHALL VIRAL INFECTION RESPONSE DN | 29 | 21 | All SZGR 2.0 genes in this pathway |
| HAHTOLA CTCL PATHOGENESIS | 16 | 9 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP | 397 | 206 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 DN | 157 | 106 | All SZGR 2.0 genes in this pathway |
| BOSCO TH1 CYTOTOXIC MODULE | 114 | 62 | All SZGR 2.0 genes in this pathway |