Gene Page: ZNF189
Summary ?
| GeneID | 7743 |
| Symbol | ZNF189 |
| Synonyms | - |
| Description | zinc finger protein 189 |
| Reference | MIM:603132|HGNC:HGNC:12980|Ensembl:ENSG00000136870|HPRD:11930|Vega:OTTHUMG00000020383 |
| Gene type | protein-coding |
| Map location | 9q22-q31 |
| Pascal p-value | 0.859 |
| Sherlock p-value | 0.001 |
| Fetal beta | 0.247 |
| eGene | Cerebellar Hemisphere Cerebellum Hippocampus Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs504023 | chr9 | 104178110 | ZNF189 | 7743 | 0.14 | cis | ||
| rs682 | chr9 | 104187019 | ZNF189 | 7743 | 0.05 | cis | ||
| rs2070396 | chr21 | 30878751 | ZNF189 | 7743 | 0.19 | trans |
Section II. Transcriptome annotation
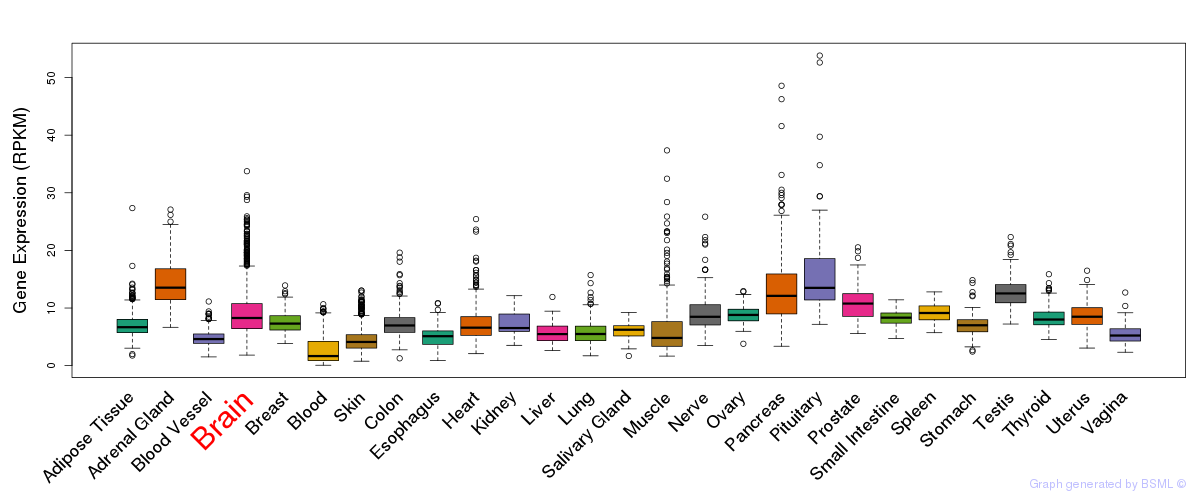
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DCAF7 | 0.94 | 0.95 |
| ZXDB | 0.94 | 0.94 |
| YTHDF2 | 0.93 | 0.93 |
| SUFU | 0.93 | 0.93 |
| GTF3C4 | 0.93 | 0.95 |
| MEX3C | 0.93 | 0.95 |
| ZNF317 | 0.93 | 0.94 |
| USP10 | 0.93 | 0.94 |
| RRP1B | 0.93 | 0.95 |
| ADNP2 | 0.93 | 0.95 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.70 | -0.86 |
| MT-CO2 | -0.70 | -0.88 |
| FXYD1 | -0.70 | -0.86 |
| C5orf53 | -0.69 | -0.75 |
| AF347015.27 | -0.69 | -0.84 |
| AF347015.33 | -0.69 | -0.84 |
| S100B | -0.69 | -0.81 |
| AIFM3 | -0.68 | -0.73 |
| MT-CYB | -0.67 | -0.84 |
| AF347015.8 | -0.67 | -0.86 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE DN | 244 | 147 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| CHEN LVAD SUPPORT OF FAILING HEART UP | 103 | 69 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| VANOEVELEN MYOGENESIS SIN3A TARGETS | 220 | 133 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 2 TRANSIENTLY INDUCED BY EGF | 51 | 29 | All SZGR 2.0 genes in this pathway |