Gene Page: CACNA1F
Summary ?
| GeneID | 778 |
| Symbol | CACNA1F |
| Synonyms | AIED|COD3|COD4|CORDX|CORDX3|CSNB2|CSNB2A|CSNBX2|Cav1.4|Cav1.4alpha1|JM8|JMC8|OA2 |
| Description | calcium voltage-gated channel subunit alpha1 F |
| Reference | MIM:300110|HGNC:HGNC:1393|Ensembl:ENSG00000102001|HPRD:02119|Vega:OTTHUMG00000022703 |
| Gene type | protein-coding |
| Map location | Xp11.23 |
| Sherlock p-value | 0.941 |
| Fetal beta | -1.305 |
| eGene | Myers' cis & trans |
| Support | EXCITABILITY SEROTONIN CompositeSet Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10949917 | chr7 | 63880876 | CACNA1F | 778 | 0.07 | trans |
Section II. Transcriptome annotation
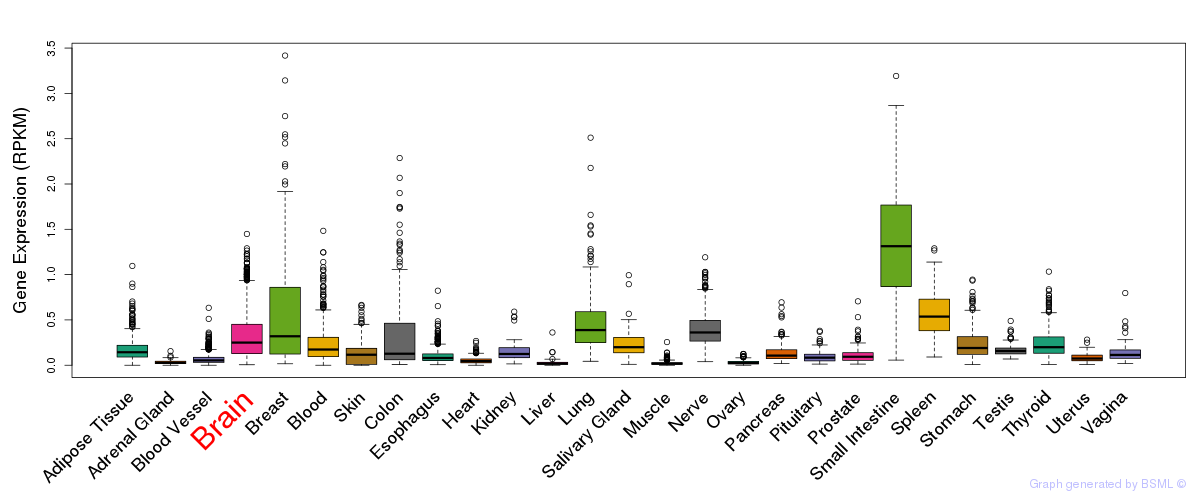
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ABR | 0.91 | 0.90 |
| STXBP1 | 0.91 | 0.90 |
| ZER1 | 0.91 | 0.89 |
| PGBD5 | 0.90 | 0.89 |
| ANKRD34A | 0.90 | 0.90 |
| PIP5K1C | 0.89 | 0.90 |
| DAGLA | 0.89 | 0.89 |
| AMIGO1 | 0.89 | 0.88 |
| ATG9A | 0.89 | 0.88 |
| PI4KA | 0.89 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.57 | -0.45 |
| GNG11 | -0.56 | -0.55 |
| AP002478.3 | -0.55 | -0.53 |
| C1orf54 | -0.53 | -0.53 |
| RAB13 | -0.52 | -0.61 |
| AF347015.18 | -0.52 | -0.42 |
| NOSTRIN | -0.52 | -0.45 |
| DBI | -0.51 | -0.56 |
| NSBP1 | -0.51 | -0.53 |
| MT-CO2 | -0.51 | -0.41 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005244 | voltage-gated ion channel activity | IEA | - | |
| GO:0005245 | voltage-gated calcium channel activity | IEA | - | |
| GO:0015270 | dihydropyridine-sensitive calcium channel activity | IDA | 15897456 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006816 | calcium ion transport | IEA | - | |
| GO:0006811 | ion transport | IEA | - | |
| GO:0050896 | response to stimulus | IEA | - | |
| GO:0050908 | detection of light stimulus involved in visual perception | IMP | 7571473 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IDA | 15897456 | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005891 | voltage-gated calcium channel complex | IDA | 15897456 | |
| GO:0005891 | voltage-gated calcium channel complex | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG CARDIAC MUSCLE CONTRACTION | 80 | 51 | All SZGR 2.0 genes in this pathway |
| KEGG VASCULAR SMOOTH MUSCLE CONTRACTION | 115 | 81 | All SZGR 2.0 genes in this pathway |
| KEGG GNRH SIGNALING PATHWAY | 101 | 77 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| KEGG HYPERTROPHIC CARDIOMYOPATHY HCM | 85 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG ARRHYTHMOGENIC RIGHT VENTRICULAR CARDIOMYOPATHY ARVC | 76 | 59 | All SZGR 2.0 genes in this pathway |
| KEGG DILATED CARDIOMYOPATHY | 92 | 68 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A12 | 317 | 177 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |