Gene Page: USP7
Summary ?
| GeneID | 7874 |
| Symbol | USP7 |
| Synonyms | HAUSP|TEF1 |
| Description | ubiquitin specific peptidase 7 (herpes virus-associated) |
| Reference | MIM:602519|HGNC:HGNC:12630|Ensembl:ENSG00000187555|HPRD:03950|Vega:OTTHUMG00000176903 |
| Gene type | protein-coding |
| Map location | 16p13.3 |
| Pascal p-value | 0.286 |
| Sherlock p-value | 0.31 |
| Fetal beta | -0.238 |
| DMG | 1 (# studies) |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.172 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| USP7 | chr16 | 9012921 | T | C | NM_003470 | . | silent | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg04444027 | 16 | 8986092 | USP7 | 5.684E-4 | 0.349 | 0.049 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
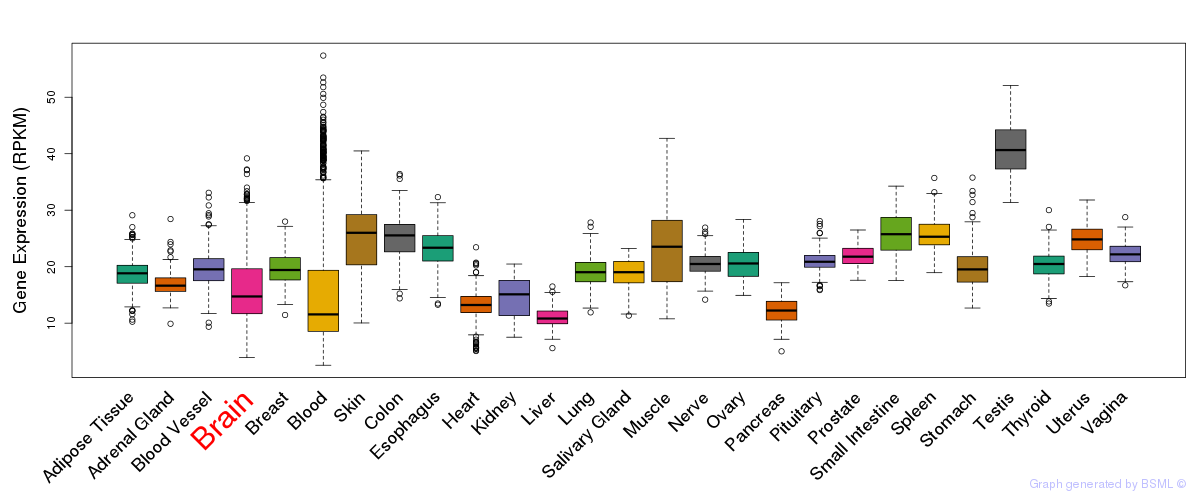
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C7orf20 | 0.90 | 0.89 |
| RMND5B | 0.89 | 0.88 |
| DPH2 | 0.88 | 0.87 |
| VAC14 | 0.87 | 0.87 |
| B4GALNT1 | 0.87 | 0.88 |
| AGPAT1 | 0.87 | 0.88 |
| RNF31 | 0.87 | 0.83 |
| RNF123 | 0.87 | 0.85 |
| DHX30 | 0.86 | 0.85 |
| CCDC22 | 0.86 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.69 | -0.68 |
| AF347015.8 | -0.68 | -0.64 |
| MT-CO2 | -0.67 | -0.62 |
| AL139819.3 | -0.67 | -0.70 |
| AF347015.31 | -0.66 | -0.62 |
| AF347015.27 | -0.66 | -0.64 |
| MT-CYB | -0.65 | -0.62 |
| AF347015.33 | -0.65 | -0.61 |
| AF347015.2 | -0.64 | -0.58 |
| AF347015.26 | -0.64 | -0.60 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004197 | cysteine-type endopeptidase activity | TAS | 9827704 | |
| GO:0004221 | ubiquitin thiolesterase activity | IEA | - | |
| GO:0008022 | protein C-terminus binding | IPI | 12093161 | |
| GO:0008234 | cysteine-type peptidase activity | IEA | - | |
| GO:0008233 | peptidase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006511 | ubiquitin-dependent protein catabolic process | IEA | - | |
| GO:0016579 | protein deubiquitination | IMP | 11923872 | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IDA | 12093161 | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATXN1 | ATX1 | D6S504E | SCA1 | ataxin 1 | - | HPRD,BioGRID | 12093161 |
| MAGEB2 | DAM6 | MAGE-XP-2 | MGC26438 | melanoma antigen family B, 2 | Affinity Capture-MS | BioGRID | 17353931 |
| MDM2 | HDMX | MGC71221 | hdm2 | Mdm2 p53 binding protein homolog (mouse) | Mdm2 interacts with HAUSP. This interaction was modeled on a demonstrated interaction between Mdm2 from an unspecified species and human HAUSP. | BIND | 15916963 |
| MDM4 | DKFZp781B1423 | HDMX | MDMX | MGC132766 | MRP1 | Mdm4 p53 binding protein homolog (mouse) | Hdmx interacts with HAUSP. | BIND | 15916963 |
| RNPS1 | E5.1 | MGC117332 | RNA binding protein S1, serine-rich domain | Affinity Capture-MS | BioGRID | 17353931 |
| RUVBL2 | CGI-46 | ECP51 | INO80J | REPTIN | RVB2 | TIH2 | TIP48 | TIP49B | RuvB-like 2 (E. coli) | Affinity Capture-MS | BioGRID | 17353931 |
| TANK | I-TRAF | TRAF2 | TRAF family member-associated NFKB activator | - | HPRD,BioGRID | 11279055 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | - | HPRD,BioGRID | 11923872 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | p53 interacts with HAUSP. | BIND | 15916963 |
| TRAF1 | EBI6 | MGC:10353 | TNF receptor-associated factor 1 | - | HPRD,BioGRID | 11279055 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | - | HPRD,BioGRID | 11279055 |
| TRAF3 | CAP-1 | CD40bp | CRAF1 | LAP1 | TNF receptor-associated factor 3 | - | HPRD,BioGRID | 11279055 |
| TRAF4 | CART1 | MLN62 | RNF83 | TNF receptor-associated factor 4 | - | HPRD,BioGRID | 11279055 |
| TRAF5 | MGC:39780 | RNF84 | TNF receptor-associated factor 5 | - | HPRD,BioGRID | 11279055 |
| TRAF6 | MGC:3310 | RNF85 | TNF receptor-associated factor 6 | - | HPRD,BioGRID | 11279055 |
| USP7 | HAUSP | TEF1 | ubiquitin specific peptidase 7 (herpes virus-associated) | - | HPRD | 12507430 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID FOXO PATHWAY | 49 | 43 | All SZGR 2.0 genes in this pathway |
| PID P53 REGULATION PATHWAY | 59 | 50 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 5 | 147 | 89 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH OCT4 TARGETS | 290 | 172 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH NOS TARGETS | 179 | 105 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION UP | 195 | 138 | All SZGR 2.0 genes in this pathway |
| STARK HYPPOCAMPUS 22Q11 DELETION UP | 53 | 40 | All SZGR 2.0 genes in this pathway |
| STARK BRAIN 22Q11 DELETION | 13 | 10 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR DN | 41 | 30 | All SZGR 2.0 genes in this pathway |
| KENNY CTNNB1 TARGETS DN | 52 | 34 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS MATURE CELL | 293 | 160 | All SZGR 2.0 genes in this pathway |
| JOSEPH RESPONSE TO SODIUM BUTYRATE DN | 64 | 45 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| AMUNDSON POOR SURVIVAL AFTER GAMMA RADIATION 2G | 171 | 96 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| GRADE COLON AND RECTAL CANCER UP | 285 | 167 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER UP | 83 | 63 | All SZGR 2.0 genes in this pathway |
| GABRIELY MIR21 TARGETS | 289 | 187 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 UP | 115 | 73 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-140 | 237 | 243 | 1A | hsa-miR-140brain | AGUGGUUUUACCCUAUGGUAG |
| miR-409-5p | 234 | 241 | 1A,m8 | hsa-miR-409-5p | AGGUUACCCGAGCAACUUUGCA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.