Gene Page: CARS2
Summary ?
| GeneID | 79587 |
| Symbol | CARS2 |
| Synonyms | COXPD27|cysRS |
| Description | cysteinyl-tRNA synthetase 2, mitochondrial (putative) |
| Reference | MIM:612800|HGNC:HGNC:25695|Ensembl:ENSG00000134905|HPRD:07763|Vega:OTTHUMG00000017347 |
| Gene type | protein-coding |
| Map location | 13q34 |
| Pascal p-value | 0.052 |
| Sherlock p-value | 0.714 |
| Fetal beta | 0.375 |
| eGene | Cortex Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
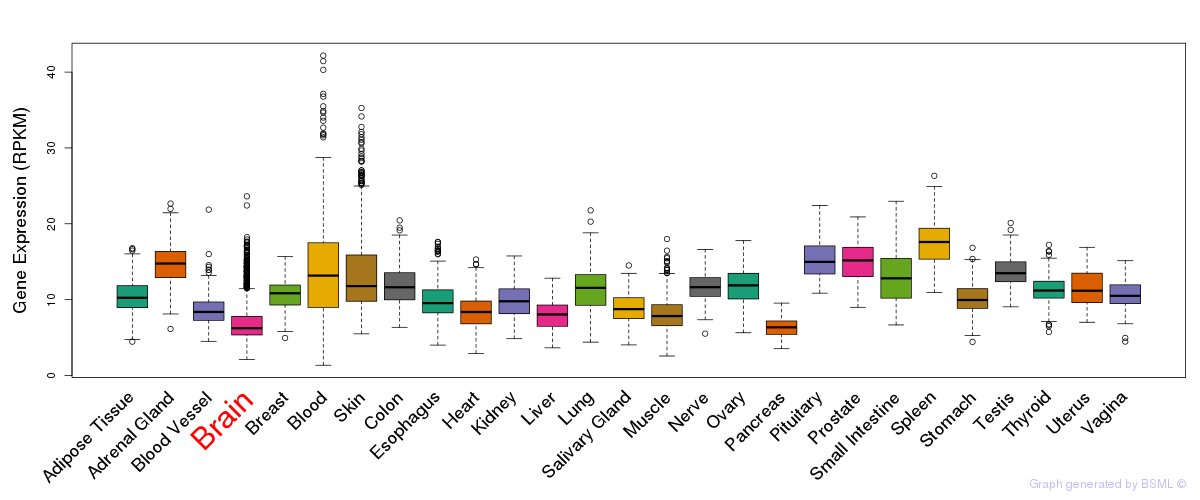
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DYDC2 | 0.94 | 0.74 |
| ZMYND10 | 0.94 | 0.69 |
| ROPN1L | 0.92 | 0.41 |
| C9orf116 | 0.92 | 0.65 |
| MORN5 | 0.92 | 0.23 |
| FAM183A | 0.92 | 0.25 |
| C6orf165 | 0.91 | 0.58 |
| TEKT1 | 0.91 | 0.49 |
| C2orf39 | 0.91 | 0.55 |
| SPATA17 | 0.91 | 0.60 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.24 | -0.44 |
| AF347015.8 | -0.24 | -0.44 |
| AF347015.2 | -0.24 | -0.47 |
| AF347015.33 | -0.23 | -0.42 |
| MT-CYB | -0.23 | -0.44 |
| AF347015.27 | -0.23 | -0.42 |
| AF347015.15 | -0.23 | -0.43 |
| CD8BP | -0.22 | -0.43 |
| AF347015.26 | -0.22 | -0.47 |
| ALDH1A1 | -0.22 | -0.38 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004817 | cysteine-tRNA ligase activity | IEA | - | |
| GO:0016874 | ligase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| GO:0046872 | metal ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006423 | cysteinyl-tRNA aminoacylation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005739 | mitochondrion | IEA | - | |
| GO:0005759 | mitochondrial matrix | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG AMINOACYL TRNA BIOSYNTHESIS | 41 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME TRNA AMINOACYLATION | 42 | 34 | All SZGR 2.0 genes in this pathway |
| VANHARANTA UTERINE FIBROID WITH 7Q DELETION UP | 67 | 37 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |