Gene Page: CALM1
Summary ?
| GeneID | 801 |
| Symbol | CALM1 |
| Synonyms | CALML2|CAMI|CPVT4|DD132|LQT14|PHKD|caM |
| Description | calmodulin 1 (phosphorylase kinase, delta) |
| Reference | MIM:114180|HGNC:HGNC:1442|Ensembl:ENSG00000198668|HPRD:00241|Vega:OTTHUMG00000171044 |
| Gene type | protein-coding |
| Map location | 14q32.11 |
| Pascal p-value | 0.044 |
| Sherlock p-value | 0.768 |
| Fetal beta | -0.689 |
| DMG | 2 (# studies) |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION SEROTONIN G2Cdb.human_mGluR5 G2Cdb.human_PocklingtonH1 G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| ADT:Sun_2012 | Systematic Investigation of Antipsychotic Drugs and Their Targets | A total of 382 drug-target associations involving 43 antipsychotic drugs and 49 target genes. | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 3 |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 3 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 14.7236 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg20328328 | 14 | 90863801 | CALM1 | 9.98E-9 | -0.01 | 4.36E-6 | DMG:Jaffe_2016 |
| cg10173240 | 14 | 90849342 | CALM1 | 3.41E-8 | -0.021 | 1.02E-5 | DMG:Jaffe_2016 |
| cg12624641 | 14 | 90420569 | CALM1 | 0.001 | 3.969 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
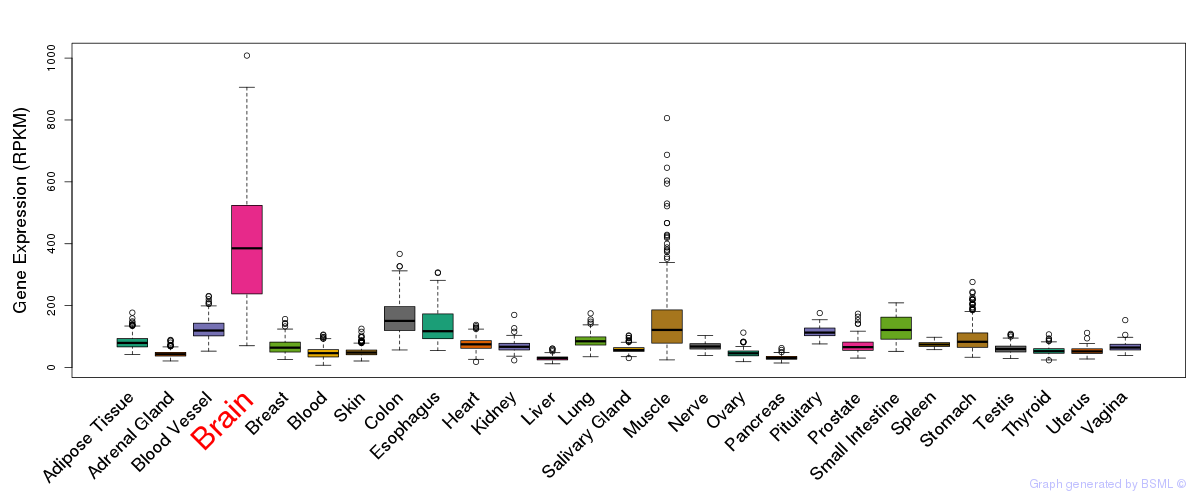
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CACYBP | 0.88 | 0.93 |
| ATP6V1D | 0.84 | 0.89 |
| SNX3 | 0.84 | 0.87 |
| NKIRAS1 | 0.83 | 0.88 |
| TCEAL7 | 0.83 | 0.92 |
| NAT5 | 0.82 | 0.88 |
| NDUFAF4 | 0.82 | 0.86 |
| NDUFA5 | 0.81 | 0.85 |
| TTC1 | 0.81 | 0.86 |
| CISD1 | 0.81 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SMTN | -0.50 | -0.55 |
| SLC27A1 | -0.49 | -0.50 |
| FAM59B | -0.48 | -0.50 |
| RP4-697K14.1 | -0.48 | -0.55 |
| PLXNB1 | -0.48 | -0.50 |
| TENC1 | -0.47 | -0.47 |
| NFKB2 | -0.47 | -0.51 |
| AC010618.2 | -0.47 | -0.50 |
| CDC42EP4 | -0.46 | -0.50 |
| SEMA4B | -0.46 | -0.46 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ADCYAP1R1 | PAC1 | PACAPR | PACAPRI | adenylate cyclase activating polypeptide 1 (pituitary) receptor type I | Calmodulin interacts with PACPR. | BIND | 15670850 |
| ADD1 | ADDA | MGC3339 | MGC44427 | adducin 1 (alpha) | Phenotypic Suppression | BioGRID | 8626479 |
| ADD1 | ADDA | MGC3339 | MGC44427 | adducin 1 (alpha) | - | HPRD | 8663089 |
| ADD2 | ADDB | adducin 2 (beta) | - | HPRD | 7490111 |
| AKAP9 | AKAP350 | AKAP450 | CG-NAP | HYPERION | KIAA0803 | MU-RMS-40.16A | PRKA9 | YOTIAO | A kinase (PRKA) anchor protein (yotiao) 9 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12221128 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Affinity Capture-Western Co-localization Reconstituted Complex | BioGRID | 14695896 |
| ASCL2 | ASH2 | HASH2 | MASH2 | bHLHa45 | achaete-scute complex homolog 2 (Drosophila) | - | HPRD | 10757985 |
| CABIN1 | CAIN | KIAA0330 | PPP3IN | calcineurin binding protein 1 | CALM1 (CaM) interacts with CABIN1. This interaction was modelled on a demonstrated interaction between CaM from an unspecified species and human CABIN1. | BIND | 15902271 |
| CALCR | CRT | CTR | CTR1 | calcitonin receptor | Calmodulin interacts with CALTR. | BIND | 15670850 |
| CALD1 | CDM | H-CAD | L-CAD | MGC21352 | NAG22 | caldesmon 1 | - | HPRD | 11736632 |
| CAMK2G | CAMK | CAMK-II | CAMKG | FLJ16043 | MGC26678 | calcium/calmodulin-dependent protein kinase II gamma | - | HPRD,BioGRID | 10860555 |
| CCNA2 | CCN1 | CCNA | cyclin A2 | Reconstituted Complex | BioGRID | 14695896 |
| CCND1 | BCL1 | D11S287E | PRAD1 | U21B31 | cyclin D1 | - | HPRD | 9837900 |
| CNN1 | SMCC | Sm-Calp | calponin 1, basic, smooth muscle | - | HPRD,BioGRID | 2161834 |2455687 |
| CRHR1 | CRF-R | CRF1 | CRFR1 | CRH-R1h | CRHR | CRHR1f | corticotropin releasing hormone receptor 1 | Calmodulin interacts with CRF1R. | BIND | 15670850 |
| EDF1 | EDF-1 | MBF1 | MGC9058 | endothelial differentiation-related factor 1 | - | HPRD | 10816571 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | EGFR interacts with calmodulin | BIND | 14960328 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | - | HPRD | 11981030 |
| ESR2 | ER-BETA | ESR-BETA | ESRB | ESTRB | Erb | NR3A2 | estrogen receptor 2 (ER beta) | - | HPRD | 11981030 |
| EWSR1 | EWS | Ewing sarcoma breakpoint region 1 | Reconstituted Complex | BioGRID | 9341188 |
| GAP43 | B-50 | PP46 | growth associated protein 43 | GAP-43 interacts with CaM. | BIND | 8872303 |
| GLP1R | MGC138331 | glucagon-like peptide 1 receptor | Calmodulin interacts with GLP1R. | BIND | 15670850 |
| GLP2R | - | glucagon-like peptide 2 receptor | Calmodulin interacts with GLP2R. | BIND | 15670850 |
| GRB7 | - | growth factor receptor-bound protein 7 | Grb7 interacts with CaM. This interaction was modeled on a demonstrated interaction between human Grb7 and CaM from rat and an unspecified species. | BIND | 15806159 |
| GRK1 | GPRK1 | RHOK | RK | G protein-coupled receptor kinase 1 | - | HPRD | 9753452 |
| GRK4 | GPRK2L | GPRK4 | GRK4a | IT11 | G protein-coupled receptor kinase 4 | Reconstituted Complex | BioGRID | 9092566 |
| GRK5 | GPRK5 | G protein-coupled receptor kinase 5 | Reconstituted Complex | BioGRID | 9092566 |
| GRM5 | GPRC1E | MGLUR5 | MGLUR5A | MGLUR5B | mGlu5 | glutamate receptor, metabotropic 5 | - | HPRD | 11953448 |
| GRM7 | FLJ40498 | GLUR7 | GPRC1G | MGLUR7 | mGlu7 | glutamate receptor, metabotropic 7 | - | HPRD | 10488094 |11953448 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | - | HPRD | 8486648 |
| IQGAP1 | HUMORFA01 | KIAA0051 | SAR1 | p195 | IQ motif containing GTPase activating protein 1 | Reconstituted Complex | BioGRID | 11734550 |12446675 |
| IQGAP2 | - | IQ motif containing GTPase activating protein 2 | - | HPRD | 8756646 |
| KCNN2 | KCa2.2 | SK2 | SKCA2 | hSK2 | potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 | - | HPRD,BioGRID | 11323678 |
| KCNQ2 | BFNC | EBN | EBN1 | ENB1 | HNSPC | KCNA11 | KV7.2 | KVEBN1 | potassium voltage-gated channel, KQT-like subfamily, member 2 | - | HPRD | 12032157 |
| KCNQ5 | Kv7.5 | potassium voltage-gated channel, KQT-like subfamily, member 5 | - | HPRD | 12032157 |
| LYST | CHS | CHS1 | lysosomal trafficking regulator | - | HPRD | 11984006 |
| MAPT | DDPAC | FLJ31424 | FTDP-17 | MAPTL | MGC138549 | MSTD | MTBT1 | MTBT2 | PPND | TAU | microtubule-associated protein tau | - | HPRD,BioGRID | 2123288 |
| MYF5 | bHLHc2 | myogenic factor 5 | - | HPRD | 10757985 |
| MYF6 | MRF4 | bHLHc4 | myogenic factor 6 (herculin) | - | HPRD | 10757985 |
| MYLK | DKFZp686I10125 | FLJ12216 | KRP | MLCK | MLCK1 | MLCK108 | MLCK210 | MSTP083 | MYLK1 | smMLCK | myosin light chain kinase | - | HPRD,BioGRID | 7947686 |
| MYO9B | CELIAC4 | MYR5 | myosin IXB | - | HPRD,BioGRID | 9490638 |
| MYOD1 | MYF3 | MYOD | PUM | bHLHc1 | myogenic differentiation 1 | - | HPRD | 10757985 |
| MYOG | MYF4 | MYOGENIN | bHLHc3 | myogenin (myogenic factor 4) | - | HPRD | 10757985 |
| NEUROD1 | BETA2 | BHF-1 | NEUROD | bHLHa3 | neurogenic differentiation 1 | - | HPRD | 10757985 |
| OPRM1 | KIAA0403 | MOR | MOR1 | OPRM | opioid receptor, mu 1 | - | HPRD,BioGRID | 11457836 |
| PCNT | KEN | MOPD2 | PCN | PCNT2 | PCNTB | PCTN2 | SCKL4 | pericentrin | Reconstituted Complex | BioGRID | 10823944 |
| PDE1A | HCAM1 | HSPDE1A | MGC26303 | phosphodiesterase 1A, calmodulin-dependent | - | HPRD,BioGRID | 12135876 |
| PHKG1 | PHKG | phosphorylase kinase, gamma 1 (muscle) | - | HPRD | 2507540 |
| PPEF1 | PP7 | PPEF | PPP7C | protein phosphatase, EF-hand calcium binding domain 1 | - | HPRD,BioGRID | 12051765 |
| PPEF2 | - | protein phosphatase, EF-hand calcium binding domain 2 | - | HPRD,BioGRID | 12051765 |
| PTH2R | PTHR2 | parathyroid hormone 2 receptor | Calmodulin interacts with PTH2R. | BIND | 15670850 |
| RAB3B | - | RAB3B, member RAS oncogene family | - | HPRD | 11741295 |
| RIT2 | RIBA | RIN | ROC2 | Ras-like without CAAX 2 | - | HPRD,BioGRID | 8824319 |
| RYR1 | CCO | MHS | MHS1 | RYDR | RYR | SKRR | ryanodine receptor 1 (skeletal) | - | HPRD | 12509414 |
| SCN4A | HYKPP | HYPP | NAC1A | Na(V)1.4 | Nav1.4 | SkM1 | sodium channel, voltage-gated, type IV, alpha subunit | Nav1.4 interacts with CaM. This interaction was modeled on a demonstrated interaction between rat Nav1.4 and human CaM. | BIND | 15746172 |
| SCTR | SR | secretin receptor | Calmodulin interacts with SECRR. | BIND | 15670850 |
| SLC9A1 | APNH | FLJ42224 | NHE1 | solute carrier family 9 (sodium/hydrogen exchanger), member 1 | Far Western | BioGRID | 12809501 |
| SNCA | MGC110988 | NACP | PARK1 | PARK4 | PD1 | synuclein, alpha (non A4 component of amyloid precursor) | - | HPRD,BioGRID | 12358748 |
| SNTA1 | SNT1 | TACIP1 | dJ1187J4.5 | syntrophin, alpha 1 (dystrophin-associated protein A1, 59kDa, acidic component) | - | HPRD | 9512352 |
| SYT1 | DKFZp781D2042 | P65 | SVP65 | SYT | synaptotagmin I | Reconstituted Complex | BioGRID | 8901523 |
| TCF3 | E2A | ITF1 | MGC129647 | MGC129648 | bHLHb21 | transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) | - | HPRD,BioGRID | 10757985 |
| TCF4 | E2-2 | ITF2 | MGC149723 | MGC149724 | PTHS | SEF2 | SEF2-1 | SEF2-1A | SEF2-1B | bHLHb19 | transcription factor 4 | - | HPRD,BioGRID | 10757985 |
| TRPV1 | DKFZp434K0220 | VR1 | transient receptor potential cation channel, subfamily V, member 1 | Affinity Capture-Western Reconstituted Complex | BioGRID | 12808128 |
| TTN | CMD1G | CMH9 | CMPD4 | CONNECTIN | DKFZp451N061 | EOMFC | FLJ26020 | FLJ26409 | FLJ32040 | FLJ34413 | FLJ39564 | FLJ43066 | HMERF | LGMD2J | TMD | titin | - | HPRD | 11178895 |
| VIPR1 | FLJ41949 | HVR1 | II | PACAP-R-2 | RDC1 | VAPC1 | VIPR | VIRG | VPAC1 | VPCAP1R | vasoactive intestinal peptide receptor 1 | Calmodulin interacts with VIP1R. | BIND | 15670850 |
| YWHAE | 14-3-3E | FLJ45465 | KCIP-1 | MDCR | MDS | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, epsilon polypeptide | - | HPRD | 10088721 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG PHOSPHATIDYLINOSITOL SIGNALING SYSTEM | 76 | 56 | All SZGR 2.0 genes in this pathway |
| KEGG OOCYTE MEIOSIS | 114 | 79 | All SZGR 2.0 genes in this pathway |
| KEGG VASCULAR SMOOTH MUSCLE CONTRACTION | 115 | 81 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM POTENTIATION | 70 | 57 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROTROPHIN SIGNALING PATHWAY | 126 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG OLFACTORY TRANSDUCTION | 389 | 85 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG GNRH SIGNALING PATHWAY | 101 | 77 | All SZGR 2.0 genes in this pathway |
| KEGG MELANOGENESIS | 102 | 80 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| KEGG GLIOMA | 65 | 56 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NO1 PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA AT1R PATHWAY | 36 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BCR PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA BIOPEPTIDES PATHWAY | 45 | 35 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CACAM PATHWAY | 16 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HDAC PATHWAY | 32 | 25 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GCR PATHWAY | 22 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CALCINEURIN PATHWAY | 21 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NDKDYNAMIN PATHWAY | 21 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FCER1 PATHWAY | 41 | 30 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FMLP PATHWAY | 39 | 29 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PYK2 PATHWAY | 31 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA VIP PATHWAY | 29 | 26 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NFAT PATHWAY | 56 | 45 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NOS1 PATHWAY | 24 | 21 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CCR5 PATHWAY | 20 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PGC1A PATHWAY | 26 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MEF2D PATHWAY | 23 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA GPCR PATHWAY | 37 | 31 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TCR PATHWAY | 49 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCOGEN BREAKDOWN GLYCOGENOLYSIS | 18 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME DAG AND IP3 SIGNALING | 32 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | 13 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB2 | 101 | 78 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY EGFR IN CANCER | 109 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | 29 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | 126 | 90 | All SZGR 2.0 genes in this pathway |
| REACTOME NGF SIGNALLING VIA TRKA FROM THE PLASMA MEMBRANE | 137 | 105 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR IN DISEASE | 127 | 88 | All SZGR 2.0 genes in this pathway |
| REACTOME ENOS ACTIVATION AND REGULATION | 20 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | 89 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMISSION ACROSS CHEMICAL SYNAPSES | 186 | 155 | All SZGR 2.0 genes in this pathway |
| REACTOME NEURONAL SYSTEM | 279 | 221 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME OPIOID SIGNALLING | 78 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME CA DEPENDENT EVENTS | 30 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME DARPP 32 EVENTS | 25 | 20 | All SZGR 2.0 genes in this pathway |
| REACTOME NEUROTRANSMITTER RECEPTOR BINDING AND DOWNSTREAM TRANSMISSION IN THE POSTSYNAPTIC CELL | 137 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME PLC BETA MEDIATED EVENTS | 43 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY PDGF | 122 | 93 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNAL TRANSDUCTION | 95 | 76 | All SZGR 2.0 genes in this pathway |
| REACTOME MUSCLE CONTRACTION | 48 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | 17 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME SMOOTH MUSCLE CONTRACTION | 25 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF NMDA RECEPTOR UPON GLUTAMATE BINDING AND POSTSYNAPTIC EVENTS | 37 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF RAS | 27 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME POST NMDA RECEPTOR ACTIVATION EVENTS | 33 | 28 | All SZGR 2.0 genes in this pathway |
| REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | 15 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM SIGNALING OF ACTIVATED FGFR | 100 | 74 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPASE C MEDIATED CASCADE | 54 | 40 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF KAINATE RECEPTORS UPON GLUTAMATE BINDING | 31 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | 11 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF CARBOHYDRATES | 247 | 154 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME GLUCOSE METABOLISM | 69 | 42 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR | 112 | 80 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES UP | 82 | 51 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA DN | 146 | 94 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA BY DMOG DN | 59 | 40 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 DN | 116 | 71 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR DN | 64 | 39 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR DN | 75 | 50 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| WEINMANN ADAPTATION TO HYPOXIA UP | 29 | 24 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| JOHANSSON BRAIN CANCER EARLY VS LATE DN | 45 | 35 | All SZGR 2.0 genes in this pathway |
| TOMLINS PROSTATE CANCER DN | 40 | 33 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN UP | 479 | 299 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY DN | 382 | 224 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 DN | 169 | 102 | All SZGR 2.0 genes in this pathway |
| PARK HSC VS MULTIPOTENT PROGENITORS DN | 18 | 12 | All SZGR 2.0 genes in this pathway |
| KUROKAWA LIVER CANCER CHEMOTHERAPY DN | 41 | 28 | All SZGR 2.0 genes in this pathway |
| IIZUKA LIVER CANCER PROGRESSION L1 G1 UP | 25 | 15 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION DN | 169 | 112 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE UP | 157 | 105 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR | 293 | 193 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| YAGI AML SURVIVAL | 129 | 87 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH3 21 DN | 49 | 29 | All SZGR 2.0 genes in this pathway |
| LU AGING BRAIN DN | 153 | 120 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS UP | 175 | 116 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 8HR UP | 105 | 73 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER DN | 540 | 340 | All SZGR 2.0 genes in this pathway |
| BERNARD PPAPDC1B TARGETS DN | 58 | 39 | All SZGR 2.0 genes in this pathway |
| HUANG DASATINIB RESISTANCE DN | 69 | 44 | All SZGR 2.0 genes in this pathway |
| FOURNIER ACINAR DEVELOPMENT LATE 2 | 277 | 172 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 17 | 181 | 101 | All SZGR 2.0 genes in this pathway |
| STAMBOLSKY TARGETS OF MUTATED TP53 UP | 49 | 35 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA CLASSICAL | 162 | 122 | All SZGR 2.0 genes in this pathway |
| GREGORY SYNTHETIC LETHAL WITH IMATINIB | 145 | 83 | All SZGR 2.0 genes in this pathway |
| THILLAINADESAN ZNF217 TARGETS UP | 44 | 22 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP | 281 | 183 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 UP | 430 | 288 | All SZGR 2.0 genes in this pathway |
| IKEDA MIR30 TARGETS UP | 116 | 87 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL DN | 146 | 88 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| let-7/98 | 472 | 478 | 1A | hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| miR-1/206 | 830 | 836 | 1A | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-129-5p | 67 | 74 | 1A,m8 | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-133 | 3463 | 3470 | 1A,m8 | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
| miR-142-3p | 1367 | 1373 | m8 | hsa-miR-142-3p | UGUAGUGUUUCCUACUUUAUGGA |
| miR-142-5p | 2900 | 2906 | 1A | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-143 | 3542 | 3548 | m8 | hsa-miR-143brain | UGAGAUGAAGCACUGUAGCUCA |
| miR-15/16/195/424/497 | 1228 | 1234 | m8 | hsa-miR-15abrain | UAGCAGCACAUAAUGGUUUGUG |
| hsa-miR-16brain | UAGCAGCACGUAAAUAUUGGCG | ||||
| hsa-miR-15bbrain | UAGCAGCACAUCAUGGUUUACA | ||||
| hsa-miR-195SZ | UAGCAGCACAGAAAUAUUGGC | ||||
| hsa-miR-424 | CAGCAGCAAUUCAUGUUUUGAA | ||||
| hsa-miR-497 | CAGCAGCACACUGUGGUUUGU | ||||
| miR-181 | 126 | 132 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-19 | 2824 | 2831 | 1A,m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-193 | 301 | 307 | m8 | hsa-miR-193a | AACUGGCCUACAAAGUCCCAG |
| hsa-miR-193b | AACUGGCCCUCAAAGUCCCGCUUU | ||||
| miR-196 | 237 | 243 | m8 | hsa-miR-196a | UAGGUAGUUUCAUGUUGUUGG |
| hsa-miR-196b | UAGGUAGUUUCCUGUUGUUGG | ||||
| miR-203.1 | 3526 | 3532 | m8 | hsa-miR-203 | UGAAAUGUUUAGGACCACUAG |
| miR-214 | 1230 | 1236 | 1A | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG | ||||
| miR-409-5p | 1552 | 1558 | 1A | hsa-miR-409-5p | AGGUUACCCGAGCAACUUUGCA |
| miR-410 | 2611 | 2617 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.