Gene Page: POLR2M
Summary ?
| GeneID | 81488 |
| Symbol | POLR2M |
| Synonyms | GCOM1|GRINL1A|Gdown|Gdown1 |
| Description | polymerase (RNA) II subunit M |
| Reference | MIM:606485|HGNC:HGNC:14862|Ensembl:ENSG00000255529|HPRD:07575|Vega:OTTHUMG00000166486 |
| Gene type | protein-coding |
| Map location | 15q21.3 |
| Pascal p-value | 0.449 |
| Sherlock p-value | 0.565 |
| eGene | Cerebellar Hemisphere Cerebellum |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
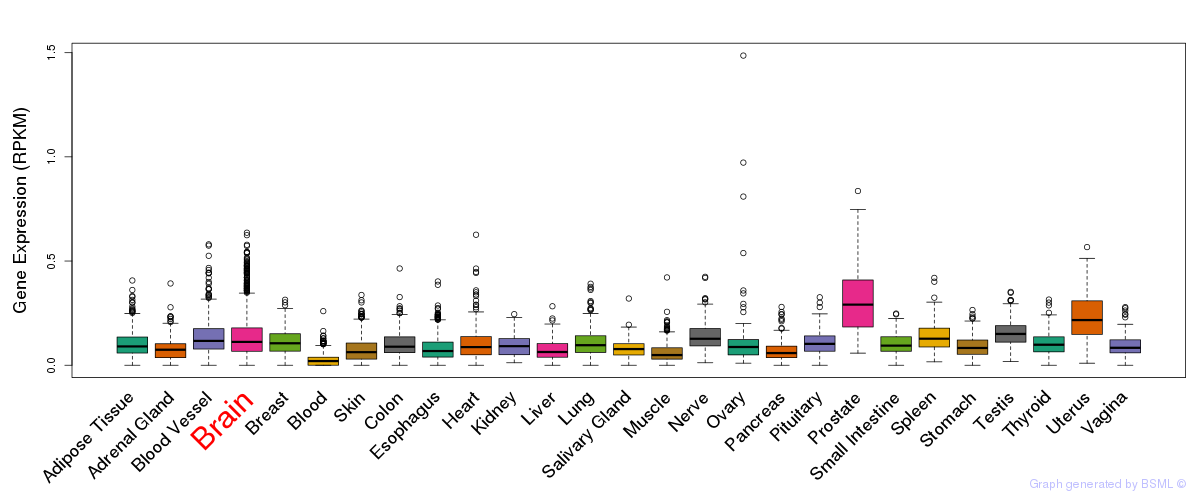
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KLHL18 | 0.87 | 0.92 |
| SLC35C1 | 0.87 | 0.92 |
| RUSC2 | 0.87 | 0.91 |
| DISP2 | 0.86 | 0.89 |
| TNPO2 | 0.85 | 0.88 |
| ZNRF1 | 0.85 | 0.89 |
| FAM123C | 0.85 | 0.88 |
| ZNF324 | 0.85 | 0.88 |
| MAPT | 0.84 | 0.90 |
| AMBRA1 | 0.84 | 0.89 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.67 | -0.76 |
| AF347015.31 | -0.67 | -0.75 |
| AF347015.33 | -0.66 | -0.74 |
| FXYD1 | -0.66 | -0.73 |
| AF347015.27 | -0.65 | -0.74 |
| AC021016.1 | -0.65 | -0.76 |
| AF347015.8 | -0.65 | -0.76 |
| MT-CYB | -0.64 | -0.73 |
| S100B | -0.64 | -0.72 |
| COPZ2 | -0.63 | -0.70 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ANKHD1 | MASK | VBARP | ankyrin repeat and KH domain containing 1 | Two-hybrid | BioGRID | 16169070 |
| APOD | - | apolipoprotein D | Two-hybrid | BioGRID | 16169070 |
| CEBPG | GPE1BP | IG/EBP-1 | CCAAT/enhancer binding protein (C/EBP), gamma | Two-hybrid | BioGRID | 16189514 |
| DGKZ | DAGK5 | DAGK6 | DGK-ZETA | hDGKzeta | diacylglycerol kinase, zeta 104kDa | Two-hybrid | BioGRID | 16169070 |
| ERAF | AHSP | EDRF | erythroid associated factor | Two-hybrid | BioGRID | 16169070 |
| HIGD1B | CLST11240 | CLST11240-15 | HIG1 domain family, member 1B | Two-hybrid | BioGRID | 16169070 |
| KIAA1377 | - | KIAA1377 | Two-hybrid | BioGRID | 16169070 |
| MYO3A | DFNB30 | myosin IIIA | Two-hybrid | BioGRID | 16169070 |
| MYO9A | FLJ11061 | FLJ13244 | MGC71859 | myosin IXA | Two-hybrid | BioGRID | 16169070 |
| PSMC2 | MGC3004 | MSS1 | Nbla10058 | S7 | proteasome (prosome, macropain) 26S subunit, ATPase, 2 | Two-hybrid | BioGRID | 16169070 |
| RBM4B | MGC10871 | RBM30 | RBM4L | ZCCHC15 | ZCRB3B | RNA binding motif protein 4B | Two-hybrid | BioGRID | 16169070 |
| RPL7A | SURF3 | TRUP | ribosomal protein L7a | Two-hybrid | BioGRID | 16169070 |
| SCMH1 | Scml3 | sex comb on midleg homolog 1 (Drosophila) | Two-hybrid | BioGRID | 16169070 |
| THAP1 | FLJ10477 | MGC33014 | THAP domain containing, apoptosis associated protein 1 | Two-hybrid | BioGRID | 16189514 |
| UQCRQ | QCR8 | QP-C | QPC | ubiquinol-cytochrome c reductase, complex III subunit VII, 9.5kDa | Two-hybrid | BioGRID | 16169070 |
| WDR89 | C14orf150 | MGC9907 | MSTP050 | WD repeat domain 89 | Two-hybrid | BioGRID | 16169070 |
| ZNF410 | APA-1 | APA1 | zinc finger protein 410 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ZHOU INFLAMMATORY RESPONSE LPS UP | 431 | 237 | All SZGR 2.0 genes in this pathway |
| CASTELLANO HRAS AND NRAS TARGETS DN | 8 | 6 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| WENDT COHESIN TARGETS UP | 33 | 19 | All SZGR 2.0 genes in this pathway |
| BOSCO ALLERGEN INDUCED TH2 ASSOCIATED MODULE | 151 | 86 | All SZGR 2.0 genes in this pathway |