Gene Page: DGCR6
Summary ?
| GeneID | 8214 |
| Symbol | DGCR6 |
| Synonyms | - |
| Description | DiGeorge syndrome critical region gene 6 |
| Reference | MIM:601279|HGNC:HGNC:2846|Ensembl:ENSG00000183628|HPRD:03177|Vega:OTTHUMG00000150162 |
| Gene type | protein-coding |
| Map location | 22q11.21|22q11 |
| Pascal p-value | 0.472 |
| Sherlock p-value | 0.205 |
| Fetal beta | -0.734 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Association | A combined odds ratio method (Sun et al. 2008), association studies | 1 | Link to SZGene |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01346830 | 22 | 18893669 | DGCR6 | 1.01E-8 | -0.017 | 4.38E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6601504 | 0 | DGCR6 | 8214 | 0.17 | trans | |||
| rs144073979 | 22 | 18886814 | DGCR6 | ENSG00000183628.8 | 4.375E-7 | 0.02 | -6727 | gtex_brain_putamen_basal |
| rs56191215 | 22 | 18887666 | DGCR6 | ENSG00000183628.8 | 3.86E-7 | 0.02 | -5875 | gtex_brain_putamen_basal |
| rs56333537 | 22 | 18888301 | DGCR6 | ENSG00000183628.8 | 1.635E-7 | 0.02 | -5240 | gtex_brain_putamen_basal |
| rs17742907 | 22 | 18890615 | DGCR6 | ENSG00000183628.8 | 1.638E-7 | 0.02 | -2926 | gtex_brain_putamen_basal |
| rs73155285 | 22 | 18890809 | DGCR6 | ENSG00000183628.8 | 1.639E-7 | 0.02 | -2732 | gtex_brain_putamen_basal |
| rs56184631 | 22 | 18891230 | DGCR6 | ENSG00000183628.8 | 1.642E-7 | 0.02 | -2311 | gtex_brain_putamen_basal |
| rs374702892 | 22 | 18894001 | DGCR6 | ENSG00000183628.8 | 2.831E-7 | 0.02 | 460 | gtex_brain_putamen_basal |
| rs55963716 | 22 | 18895875 | DGCR6 | ENSG00000183628.8 | 1.673E-7 | 0.02 | 2334 | gtex_brain_putamen_basal |
| rs73155293 | 22 | 18896087 | DGCR6 | ENSG00000183628.8 | 1.588E-7 | 0.02 | 2546 | gtex_brain_putamen_basal |
| rs10602 | 22 | 18899484 | DGCR6 | ENSG00000183628.8 | 1.093E-7 | 0.02 | 5943 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
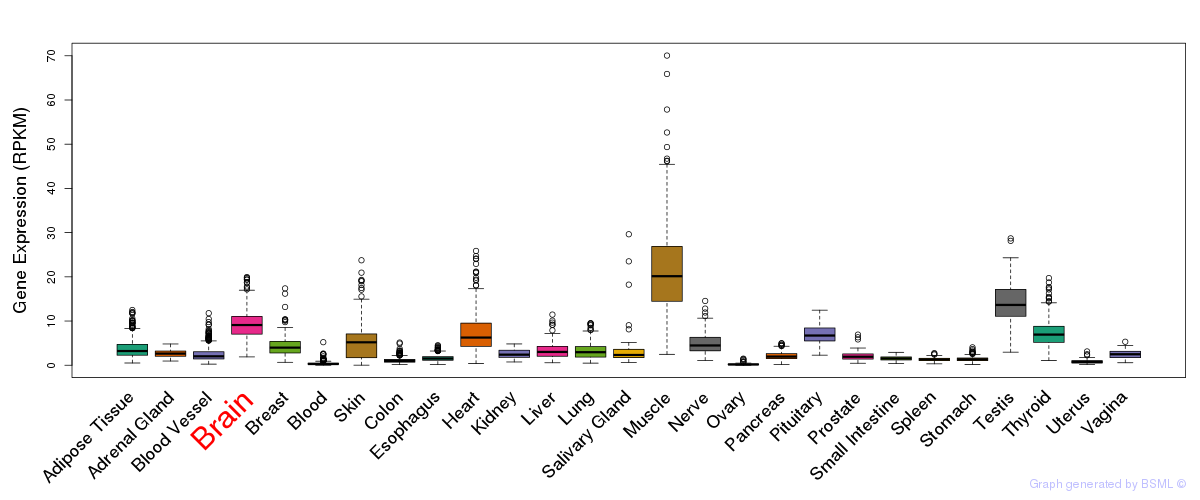
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003674 | molecular_function | ND | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007155 | cell adhesion | TAS | 8733130 | |
| GO:0009887 | organ morphogenesis | TAS | 8733130 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005578 | proteinaceous extracellular matrix | TAS | 8733130 | |
| GO:0005634 | nucleus | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| CHIARADONNA NEOPLASTIC TRANSFORMATION CDC25 DN | 153 | 100 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G DN | 35 | 18 | All SZGR 2.0 genes in this pathway |
| TERAMOTO OPN TARGETS CLUSTER 4 | 13 | 8 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| STARK PREFRONTAL CORTEX 22Q11 DELETION DN | 517 | 309 | All SZGR 2.0 genes in this pathway |
| STARK HYPPOCAMPUS 22Q11 DELETION DN | 20 | 19 | All SZGR 2.0 genes in this pathway |
| MORI MATURE B LYMPHOCYTE UP | 90 | 62 | All SZGR 2.0 genes in this pathway |
| MORI MATURE B LYMPHOCYTE DN | 75 | 43 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK UP | 244 | 151 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 UP | 428 | 266 | All SZGR 2.0 genes in this pathway |
| ABE INNER EAR | 48 | 26 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS UP | 374 | 247 | All SZGR 2.0 genes in this pathway |
| WALLACE JAK2 TARGETS UP | 26 | 15 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 13 | 172 | 107 | All SZGR 2.0 genes in this pathway |
| LI DCP2 BOUND MRNA | 89 | 57 | All SZGR 2.0 genes in this pathway |
| BAKKER FOXO3 TARGETS DN | 187 | 109 | All SZGR 2.0 genes in this pathway |