Gene Page: UXT
Summary ?
| GeneID | 8409 |
| Symbol | UXT |
| Synonyms | ART-27|STAP1 |
| Description | ubiquitously expressed prefoldin like chaperone |
| Reference | MIM:300234|HGNC:HGNC:12641|Ensembl:ENSG00000126756|HPRD:02209|Vega:OTTHUMG00000021453 |
| Gene type | protein-coding |
| Map location | Xp11.23-p11.22 |
| Sherlock p-value | 0.289 |
| Fetal beta | 0.931 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
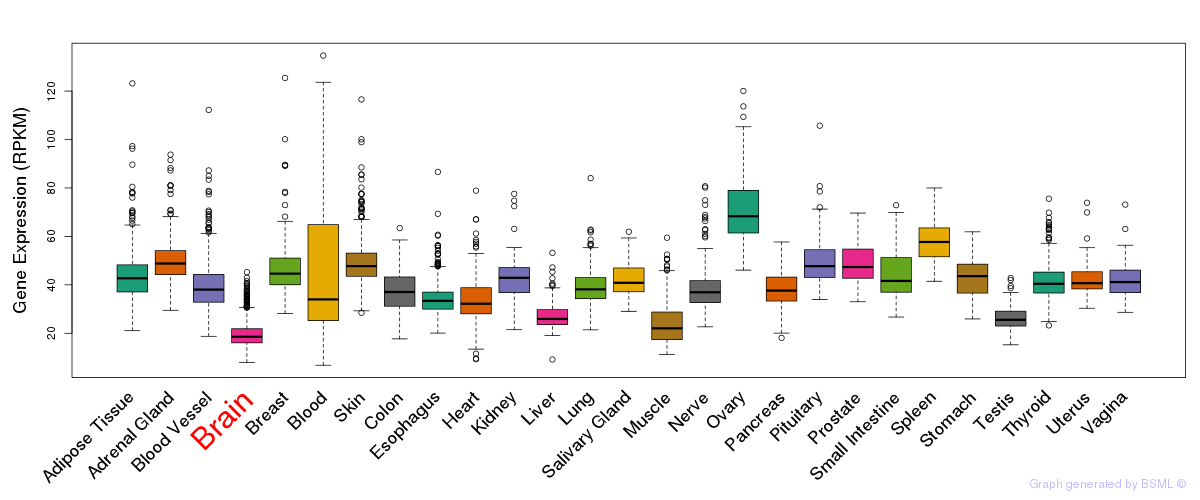
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC009163.1 | 0.73 | 0.54 |
| C1orf192 | 0.72 | 0.60 |
| FBXL13 | 0.71 | 0.48 |
| C20orf26 | 0.71 | 0.65 |
| TMEM47 | 0.70 | 0.70 |
| CRISPLD1 | 0.70 | 0.56 |
| UBXN10 | 0.69 | 0.59 |
| CCDC103 | 0.69 | 0.54 |
| IQCH | 0.69 | 0.50 |
| MDFIC | 0.68 | 0.59 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.25 | -0.30 |
| CXCL14 | -0.24 | -0.22 |
| AF347015.2 | -0.23 | -0.25 |
| AF347015.31 | -0.23 | -0.27 |
| AP000679.1 | -0.23 | -0.29 |
| AC098691.2 | -0.23 | -0.33 |
| CSAG1 | -0.22 | -0.28 |
| AF347015.21 | -0.22 | -0.28 |
| PLA2G5 | -0.22 | -0.21 |
| AF347015.8 | -0.22 | -0.26 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0008017 | microtubule binding | TAS | 12762840 | |
| GO:0051082 | unfolded protein binding | IEA | - | |
| GO:0051015 | actin filament binding | IDA | 12762840 | |
| GO:0048487 | beta-tubulin binding | IDA | 12762840 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000226 | microtubule cytoskeleton organization | IMP | 16221885 | |
| GO:0006457 | protein folding | IEA | - | |
| GO:0051297 | centrosome organization | IMP | 16221885 | |
| GO:0047497 | mitochondrion transport along microtubule | TAS | 12762840 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000930 | gamma-tubulin complex | IDA | 16221885 | |
| GO:0005813 | centrosome | IDA | 16221885 | |
| GO:0005856 | cytoskeleton | IDA | 12762840 | |
| GO:0005634 | nucleus | IDA | 12762840 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0016272 | prefoldin complex | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | - | HPRD,BioGRID | 11854421 |
| BUB3 | BUB3L | hBUB3 | budding uninhibited by benzimidazoles 3 homolog (yeast) | Two-hybrid | BioGRID | 12762840 |
| C19orf2 | FLJ10575 | NNX3 | RMP | URI | chromosome 19 open reading frame 2 | URI interacts with STAP1. | BIND | 14615539 |
| CECR2 | KIAA1740 | cat eye syndrome chromosome region, candidate 2 | - | HPRD,BioGRID | 12762840 |
| CITED2 | MRG1 | P35SRJ | Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2 | Two-hybrid | BioGRID | 12762840 |
| E2F1 | E2F-1 | RBAP1 | RBBP3 | RBP3 | E2F transcription factor 1 | E2F1 interacts with the UXT chromatin. | BIND | 11799066 |
| E2F2 | E2F-2 | E2F transcription factor 2 | E2F2 interacts with the UXT chromatin. | BIND | 11799066 |
| E2F3 | DKFZp686C18211 | E2F-3 | KIAA0075 | MGC104598 | E2F transcription factor 3 | E2F3 interacts with the UXT chromatin. | BIND | 11799066 |
| E2F4 | E2F-4 | E2F transcription factor 4, p107/p130-binding | E2F4 interacts with the UXT chromatin. | BIND | 11799066 |
| GLE1 | GLE1L | LCCS | LCCS1 | hGLE1 | GLE1 RNA export mediator homolog (yeast) | Two-hybrid | BioGRID | 14645504 |
| LRPPRC | CLONE-23970 | GP130 | LRP130 | LSFC | leucine-rich PPR-motif containing | - | HPRD,BioGRID | 11827465 |
| LSM1 | CASM | YJL124C | LSM1 homolog, U6 small nuclear RNA associated (S. cerevisiae) | - | HPRD,BioGRID | 15231747 |
| RBL1 | CP107 | MGC40006 | PRB1 | p107 | retinoblastoma-like 1 (p107) | p107 interacts with the UXT chromatin. | BIND | 11799066 |
| RBL2 | FLJ26459 | P130 | Rb2 | retinoblastoma-like 2 (p130) | p130 interacts with the UXT chromatin. | BIND | 11799066 |
| SKP2 | FBL1 | FBXL1 | FLB1 | MGC1366 | S-phase kinase-associated protein 2 (p45) | STAP1 interacts with SKP2. | BIND | 14615539 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID E2F PATHWAY | 74 | 48 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 DN | 126 | 86 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 2 DN | 77 | 46 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 DN | 162 | 116 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC UP | 72 | 53 | All SZGR 2.0 genes in this pathway |
| DANG MYC TARGETS UP | 143 | 100 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |