Gene Page: SCEL
Summary ?
| GeneID | 8796 |
| Symbol | SCEL |
| Synonyms | - |
| Description | sciellin |
| Reference | MIM:604112|HGNC:HGNC:10573|Ensembl:ENSG00000136155|HPRD:09161|Vega:OTTHUMG00000017107 |
| Gene type | protein-coding |
| Map location | 13q22 |
| Pascal p-value | 0.151 |
| Fetal beta | -0.08 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
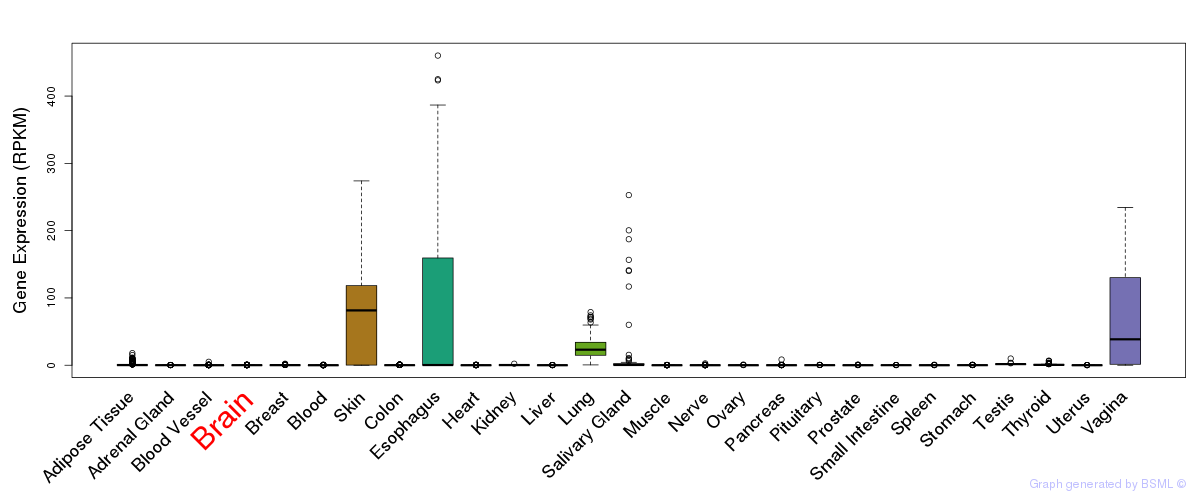
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LYZ | 0.78 | 0.62 |
| SELL | 0.73 | 0.59 |
| FCGR3B | 0.73 | 0.44 |
| S100A9 | 0.72 | 0.53 |
| OLFM4 | 0.69 | 0.38 |
| S100A8 | 0.68 | 0.47 |
| S100A12 | 0.66 | 0.45 |
| ELANE | 0.66 | 0.49 |
| SQRDL | 0.56 | 0.55 |
| FCN1 | 0.55 | 0.49 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CEP250 | -0.28 | -0.38 |
| CCDC12 | -0.27 | -0.46 |
| CEP164 | -0.27 | -0.39 |
| ZNF417 | -0.27 | -0.41 |
| UPF3A | -0.27 | -0.42 |
| HIC2 | -0.26 | -0.42 |
| ZC3H13 | -0.26 | -0.38 |
| TOP1MT | -0.26 | -0.47 |
| C14orf102 | -0.26 | -0.41 |
| PFDN2 | -0.26 | -0.46 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 DN | 175 | 82 | All SZGR 2.0 genes in this pathway |
| BARRIER COLON CANCER RECURRENCE UP | 42 | 28 | All SZGR 2.0 genes in this pathway |
| JAEGER METASTASIS DN | 258 | 141 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS DN | 232 | 139 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC2 TARGETS DN | 133 | 77 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| COLDREN GEFITINIB RESISTANCE DN | 230 | 115 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| AIGNER ZEB1 TARGETS | 35 | 15 | All SZGR 2.0 genes in this pathway |
| RICKMAN HEAD AND NECK CANCER E | 89 | 44 | All SZGR 2.0 genes in this pathway |
| LIN SILENCED BY TUMOR MICROENVIRONMENT | 108 | 73 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 DN | 169 | 102 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 3 DN | 59 | 32 | All SZGR 2.0 genes in this pathway |
| HUANG FOXA2 TARGETS DN | 36 | 21 | All SZGR 2.0 genes in this pathway |
| CROMER TUMORIGENESIS DN | 51 | 29 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS DN | 435 | 289 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS KERATINOCYTE DN | 23 | 15 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A4 | 196 | 124 | All SZGR 2.0 genes in this pathway |
| FONTAINE FOLLICULAR THYROID ADENOMA DN | 68 | 45 | All SZGR 2.0 genes in this pathway |
| FONTAINE PAPILLARY THYROID CARCINOMA UP | 66 | 38 | All SZGR 2.0 genes in this pathway |
| ONGUSAHA BRCA1 TARGETS DN | 14 | 9 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR DN | 88 | 59 | All SZGR 2.0 genes in this pathway |
| WANG MLL TARGETS | 289 | 188 | All SZGR 2.0 genes in this pathway |
| BOSCO EPITHELIAL DIFFERENTIATION MODULE | 69 | 31 | All SZGR 2.0 genes in this pathway |