Gene Page: EIF2B5
Summary ?
| GeneID | 8893 |
| Symbol | EIF2B5 |
| Synonyms | CACH|CLE|EIF-2B|EIF2Bepsilon|LVWM |
| Description | eukaryotic translation initiation factor 2B subunit epsilon |
| Reference | MIM:603945|HGNC:HGNC:3261|Ensembl:ENSG00000145191|HPRD:04898|Vega:OTTHUMG00000156840 |
| Gene type | protein-coding |
| Map location | 3q27.1 |
| Pascal p-value | 0.276 |
| Sherlock p-value | 0.855 |
| Fetal beta | 0.133 |
| DMG | 2 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg25739137 | 3 | 183851986 | EIF2B5 | 6.05E-6 | -0.357 | 0.011 | DMG:Wockner_2014 |
| cg18313702 | 3 | 183853126 | EIF2B5 | 4.02E-8 | -0.008 | 1.13E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
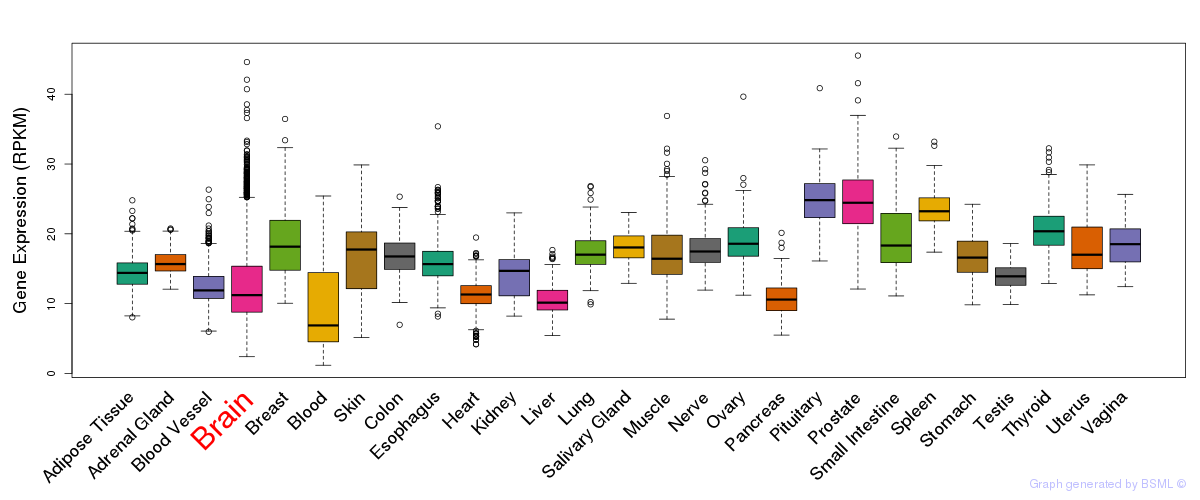
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005085 | guanyl-nucleotide exchange factor activity | IDA | 11323413 | |
| GO:0005085 | guanyl-nucleotide exchange factor activity | IMP | 15054402 | |
| GO:0005085 | guanyl-nucleotide exchange factor activity | ISS | - | |
| GO:0003743 | translation initiation factor activity | IDA | 16289705 | |
| GO:0003743 | translation initiation factor activity | NAS | 8688466 | |
| GO:0005515 | protein binding | IPI | 15060152 | |
| GO:0005515 | protein binding | ISS | - | |
| GO:0016740 | transferase activity | IEA | - | |
| GO:0008415 | acyltransferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0014003 | oligodendrocyte development | IMP | axon, oligodendrocyte, Glial (GO term level: 10) | 15217090 |
| GO:0014002 | astrocyte development | IMP | neuron, astrocyte, Glial (GO term level: 10) | 15723074 |
| GO:0042552 | myelination | IMP | neuron, axon, Brain, oligodendrocyte (GO term level: 13) | 14566705 |15723074 |
| GO:0001541 | ovarian follicle development | IMP | 15507143 | |
| GO:0009408 | response to heat | IMP | 15723074 | |
| GO:0009408 | response to heat | ISS | - | |
| GO:0009408 | response to heat | TAS | 12499492 | |
| GO:0009749 | response to glucose stimulus | ISS | - | |
| GO:0043434 | response to peptide hormone stimulus | ISS | - | |
| GO:0045948 | positive regulation of translational initiation | ISS | - | |
| GO:0051716 | cellular response to stimulus | IDA | 8626696 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005851 | eukaryotic translation initiation factor 2B complex | IDA | 11323413 |15060152 | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IDA | 11323413 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA EIF2 PATHWAY | 11 | 8 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IGF1MTOR PATHWAY | 20 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA VEGF PATHWAY | 29 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSLATION | 222 | 75 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA DN | 526 | 357 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| SLEBOS HEAD AND NECK CANCER WITH HPV UP | 84 | 43 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP UP | 265 | 158 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PENG GLUTAMINE DEPRIVATION DN | 337 | 230 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION UP | 314 | 201 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM UP | 84 | 58 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| DAIRKEE CANCER PRONE RESPONSE BPA E2 | 118 | 65 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 14 | 143 | 86 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-200bc/429 | 136 | 142 | m8 | hsa-miR-200b | UAAUACUGCCUGGUAAUGAUGAC |
| hsa-miR-200c | UAAUACUGCCGGGUAAUGAUGG | ||||
| hsa-miR-429 | UAAUACUGUCUGGUAAAACCGU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.