Gene Page: P2RX6
Summary ?
| GeneID | 9127 |
| Symbol | P2RX6 |
| Synonyms | P2RXL1|P2X6|P2XM |
| Description | purinergic receptor P2X 6 |
| Reference | MIM:608077|HGNC:HGNC:8538|HPRD:08490| |
| Gene type | protein-coding |
| Map location | 22q11.21 |
| Pascal p-value | 0.334 |
| DMG | 1 (# studies) |
| eGene | Cortex Meta |
| Support | GPCR SIGNALLING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01038149 | 22 | 21368765 | P2RX6 | 1.36E-10 | -0.01 | 4.9E-7 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
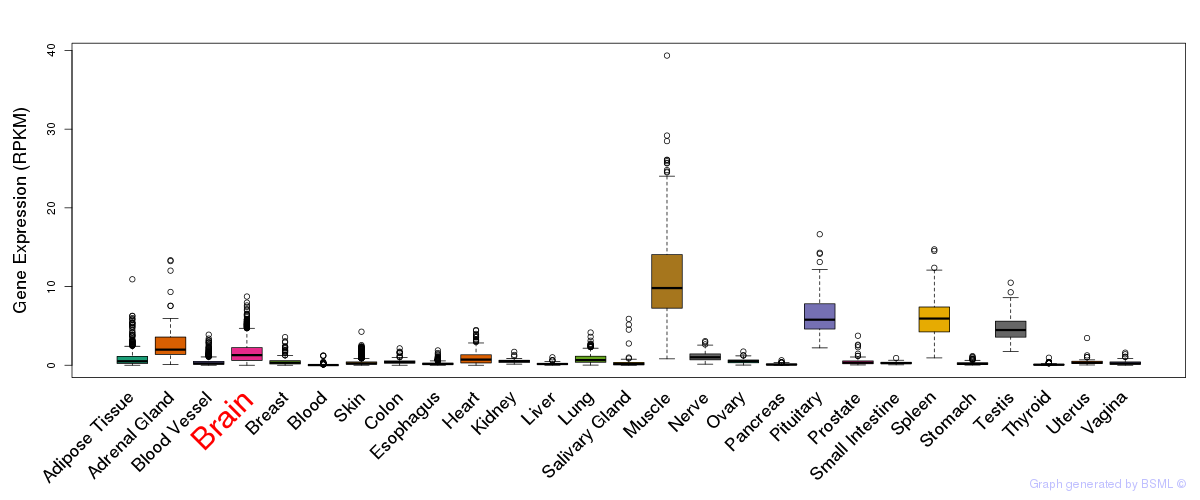
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ALK | 0.79 | 0.70 |
| ABLIM3 | 0.79 | 0.77 |
| ATBF1 | 0.75 | 0.69 |
| PEG10 | 0.74 | 0.73 |
| AC008522.1 | 0.74 | 0.74 |
| IGSF9B | 0.74 | 0.75 |
| CDR2 | 0.74 | 0.83 |
| RGMB | 0.73 | 0.74 |
| KIAA1024 | 0.73 | 0.81 |
| BLCAP | 0.73 | 0.72 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.50 | -0.62 |
| MT-CO2 | -0.48 | -0.60 |
| AF347015.21 | -0.48 | -0.63 |
| COPZ2 | -0.46 | -0.55 |
| HIGD1B | -0.46 | -0.58 |
| AF347015.33 | -0.45 | -0.53 |
| MT-CYB | -0.45 | -0.55 |
| AF347015.8 | -0.45 | -0.57 |
| AF347015.2 | -0.45 | -0.53 |
| LGALS3 | -0.43 | -0.60 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0001614 | purinergic nucleotide receptor activity | NAS | 12088286 | |
| GO:0004888 | transmembrane receptor activity | TAS | 9242461 | |
| GO:0004872 | receptor activity | IEA | - | |
| GO:0004931 | ATP-gated cation channel activity | NAS | 12088286 | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0005216 | ion channel activity | IEA | - | |
| GO:0015267 | channel activity | TAS | 9242461 | |
| GO:0046982 | protein heterodimerization activity | NAS | 17895406 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007165 | signal transduction | TAS | 9242461 | |
| GO:0006811 | ion transport | IEA | - | |
| GO:0006936 | muscle contraction | TAS | 9242461 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IDA | 12088286 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 9242461 | |
| GO:0030054 | cell junction | IDA | 12088286 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CALCIUM SIGNALING PATHWAY | 178 | 134 | All SZGR 2.0 genes in this pathway |
| KEGG NEUROACTIVE LIGAND RECEPTOR INTERACTION | 272 | 195 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS G UP | 238 | 135 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING DN | 35 | 24 | All SZGR 2.0 genes in this pathway |
| LEIN PONS MARKERS | 89 | 59 | All SZGR 2.0 genes in this pathway |
| LEIN MEDULLA MARKERS | 81 | 48 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF LCP WITH H3K4ME3 | 128 | 68 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS LCP WITH H3K4ME3 | 174 | 100 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES LCP WITH H3K4ME3 | 142 | 80 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| CERIBELLI GENES INACTIVE AND BOUND BY NFY | 45 | 27 | All SZGR 2.0 genes in this pathway |
| ACEVEDO FGFR1 TARGETS IN PROSTATE CANCER MODEL DN | 308 | 187 | All SZGR 2.0 genes in this pathway |