Gene Page: NEMF
Summary ?
| GeneID | 9147 |
| Symbol | NEMF |
| Synonyms | NY-CO-1|SDCCAG1 |
| Description | nuclear export mediator factor |
| Reference | MIM:608378|HGNC:HGNC:10663|HPRD:08494| |
| Gene type | protein-coding |
| Map location | 14q22 |
| Pascal p-value | 0.003 |
| Fetal beta | 0.388 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 7 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15453599 | 14 | 50155052 | NEMF | 1.55E-10 | -0.019 | 5E-7 | DMG:Jaffe_2016 |
| cg16537350 | 14 | 50329699 | NEMF | 2.41E-9 | -0.01 | 1.81E-6 | DMG:Jaffe_2016 |
| cg15522118 | 14 | 50155002 | NEMF | 2.49E-8 | -0.018 | 8.02E-6 | DMG:Jaffe_2016 |
| cg06798092 | 14 | 50328478 | NEMF | 2.71E-8 | -0.014 | 8.56E-6 | DMG:Jaffe_2016 |
| cg01264297 | 14 | 50234605 | NEMF | 3.22E-8 | -0.009 | 9.71E-6 | DMG:Jaffe_2016 |
| cg15365950 | 14 | 50160307 | NEMF | 5.07E-8 | -0.011 | 1.35E-5 | DMG:Jaffe_2016 |
| cg12793744 | 14 | 50101962 | NEMF | 7.19E-8 | -0.006 | 1.73E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17034560 | chr1 | 10229157 | NEMF | 9147 | 0.19 | trans | ||
| rs520325 | chr3 | 165361016 | NEMF | 9147 | 0.12 | trans | ||
| rs830202 | chr3 | 165654796 | NEMF | 9147 | 0.15 | trans | ||
| rs7076439 | chr10 | 93286841 | NEMF | 9147 | 0.16 | trans |
Section II. Transcriptome annotation
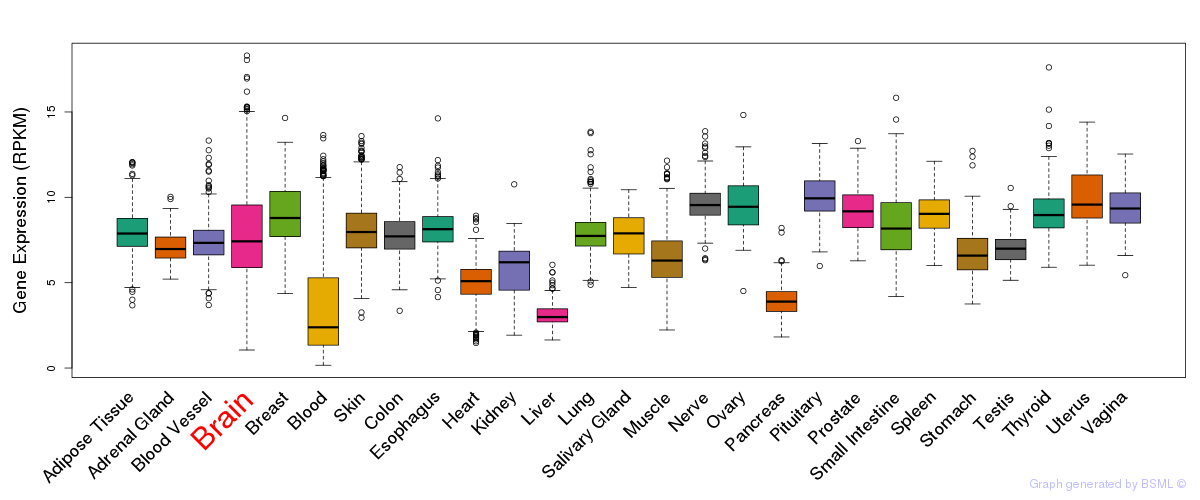
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RIC8A | 0.93 | 0.95 |
| GFOD2 | 0.92 | 0.94 |
| RAB35 | 0.92 | 0.94 |
| COBRA1 | 0.92 | 0.93 |
| KLHL22 | 0.92 | 0.94 |
| PRMT2 | 0.91 | 0.92 |
| SNX15 | 0.91 | 0.94 |
| PSMD3 | 0.91 | 0.92 |
| SH3BP5L | 0.91 | 0.92 |
| BYSL | 0.91 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.79 | -0.86 |
| MT-CO2 | -0.79 | -0.86 |
| AF347015.27 | -0.78 | -0.87 |
| AF347015.33 | -0.78 | -0.85 |
| MT-CYB | -0.77 | -0.85 |
| AF347015.8 | -0.76 | -0.86 |
| AF347015.15 | -0.73 | -0.84 |
| S100B | -0.73 | -0.80 |
| FXYD1 | -0.72 | -0.81 |
| C5orf53 | -0.72 | -0.74 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS C UP | 170 | 114 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING VIA SMAD4 DN | 66 | 38 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| ASTIER INTEGRIN SIGNALING | 59 | 44 | All SZGR 2.0 genes in this pathway |
| BILD E2F3 ONCOGENIC SIGNATURE | 246 | 153 | All SZGR 2.0 genes in this pathway |
| BILD CTNNB1 ONCOGENIC SIGNATURE | 82 | 52 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE UP | 393 | 244 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| CHEN HOXA5 TARGETS 9HR UP | 223 | 132 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS UP | 518 | 299 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 DN | 92 | 60 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL DN | 146 | 88 | All SZGR 2.0 genes in this pathway |