Gene Page: ZW10
Summary ?
| GeneID | 9183 |
| Symbol | ZW10 |
| Synonyms | HZW10|KNTC1AP |
| Description | zw10 kinetochore protein |
| Reference | MIM:603954|HGNC:HGNC:13194|Ensembl:ENSG00000086827|HPRD:04902|Vega:OTTHUMG00000168190 |
| Gene type | protein-coding |
| Map location | 11q23.2 |
| Pascal p-value | 0.221 |
| Sherlock p-value | 0.248 |
| Fetal beta | 0.197 |
| DMG | 1 (# studies) |
| eGene | Nucleus accumbens basal ganglia |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.006 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg00295835 | 11 | 113644563 | ZW10 | 6.98E-9 | -0.012 | 3.55E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
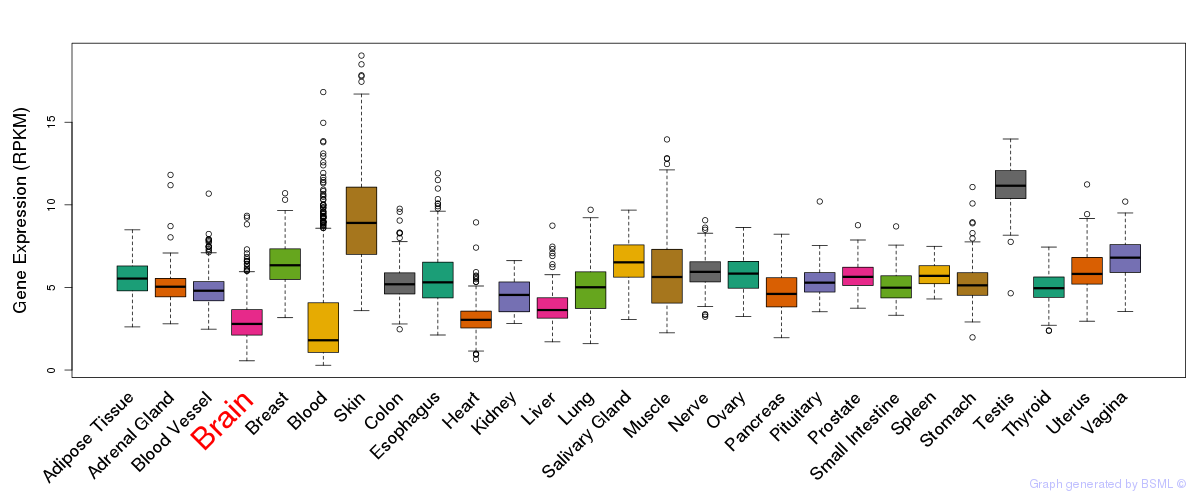
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LRMP | 0.62 | 0.65 |
| KLK7 | 0.55 | 0.54 |
| C13orf36 | 0.54 | 0.64 |
| CCDC28A | 0.52 | 0.54 |
| POPDC3 | 0.52 | 0.63 |
| HPRT1 | 0.51 | 0.54 |
| AC090772.4 | 0.50 | 0.51 |
| MAPK3 | 0.49 | 0.54 |
| AC008670.2 | 0.49 | 0.35 |
| TSPAN16 | 0.49 | 0.32 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HYAL2 | -0.38 | -0.48 |
| TBC1D16 | -0.38 | -0.43 |
| TES | -0.38 | -0.50 |
| SMTN | -0.37 | -0.46 |
| SH3BP2 | -0.36 | -0.51 |
| GLIS2 | -0.36 | -0.44 |
| CBX8 | -0.36 | -0.35 |
| FLT4 | -0.36 | -0.42 |
| SBNO2 | -0.36 | -0.42 |
| LRP5 | -0.36 | -0.45 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 11146660 |12686595 | |
| GO:0019237 | centromeric DNA binding | TAS | 9700164 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000070 | mitotic sister chromatid segregation | IDA | 11146660 | |
| GO:0006461 | protein complex assembly | IDA | 11146660 | |
| GO:0007049 | cell cycle | IEA | - | |
| GO:0007093 | mitotic cell cycle checkpoint | IDA | 11146660 | |
| GO:0007096 | regulation of exit from mitosis | IDA | 11146660 | |
| GO:0007126 | meiosis | TAS | 9298984 | |
| GO:0006888 | ER to Golgi vesicle-mediated transport | IMP | 15029241 | |
| GO:0016192 | vesicle-mediated transport | IEA | - | |
| GO:0015031 | protein transport | IEA | - | |
| GO:0051301 | cell division | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0000777 | condensed chromosome kinetochore | IEA | - | |
| GO:0000922 | spindle pole | IDA | 11146660 | |
| GO:0005828 | kinetochore microtubule | IDA | 11146660 | |
| GO:0005789 | endoplasmic reticulum membrane | IEA | - | |
| GO:0005634 | nucleus | IDA | 15485811 | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005783 | endoplasmic reticulum | IDA | 15029241 | |
| GO:0016020 | membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC PROMETAPHASE | 87 | 51 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP | 178 | 111 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT DN | 165 | 106 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| BREDEMEYER RAG SIGNALING NOT VIA ATM DN | 57 | 34 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS UP | 295 | 155 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |