Gene Page: MTA2
Summary ?
| GeneID | 9219 |
| Symbol | MTA2 |
| Synonyms | MTA1L1|PID |
| Description | metastasis associated 1 family member 2 |
| Reference | MIM:603947|HGNC:HGNC:7411|Ensembl:ENSG00000149480|HPRD:07233|Vega:OTTHUMG00000167684 |
| Gene type | protein-coding |
| Map location | 11q12-q13.1 |
| Pascal p-value | 0.454 |
| Sherlock p-value | 0.183 |
| DEG p-value | DEG:Zhao_2015:p=4.40e-04:q=0.0926 |
| eGene | Myers' cis & trans |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DEG:Zhao_2015 | RNA Sequencing analysis | Transcriptome sequencing and genome-wide association analyses reveal lysosomal function and actin cytoskeleton remodeling in schizophrenia and bipolar disorder. |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1548929 | chr16 | 49017307 | MTA2 | 9219 | 0.19 | trans | ||
| rs11872835 | chr18 | 59420762 | MTA2 | 9219 | 0.1 | trans | ||
| rs2296236 | chr20 | 3020089 | MTA2 | 9219 | 0.01 | trans | ||
| rs6138990 | chr20 | 3022617 | MTA2 | 9219 | 0.1 | trans | ||
| rs3761244 | chr20 | 3023180 | MTA2 | 9219 | 0.1 | trans |
Section II. Transcriptome annotation
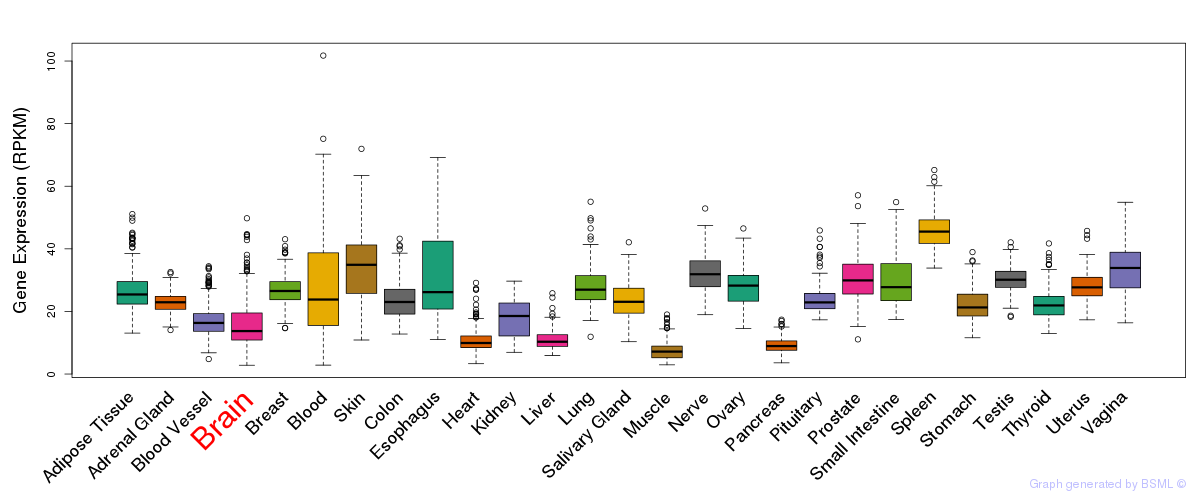
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GAPVD1 | 0.95 | 0.95 |
| SEL1L | 0.94 | 0.95 |
| ADAR | 0.94 | 0.95 |
| ZBTB38 | 0.93 | 0.95 |
| ANAPC1 | 0.93 | 0.94 |
| KIAA1219 | 0.93 | 0.94 |
| ATRN | 0.93 | 0.95 |
| HECTD1 | 0.93 | 0.93 |
| KPNA6 | 0.92 | 0.94 |
| EXOC6B | 0.92 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.70 | -0.71 |
| AF347015.21 | -0.69 | -0.74 |
| MT-CO2 | -0.69 | -0.71 |
| HIGD1B | -0.68 | -0.72 |
| FXYD1 | -0.68 | -0.69 |
| IFI27 | -0.66 | -0.69 |
| AF347015.8 | -0.66 | -0.69 |
| C1orf54 | -0.65 | -0.75 |
| CST3 | -0.65 | -0.68 |
| CXCL14 | -0.64 | -0.67 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATR | FRP1 | MEC1 | SCKL | SCKL1 | ataxia telangiectasia and Rad3 related | Affinity Capture-Western | BioGRID | 10545197 |
| CHD3 | Mi-2a | Mi2-ALPHA | ZFH | chromodomain helicase DNA binding protein 3 | Co-purification | BioGRID | 10444591 |
| CHD4 | DKFZp686E06161 | Mi-2b | Mi2-BETA | chromodomain helicase DNA binding protein 4 | Affinity Capture-MS Affinity Capture-Western | BioGRID | 12920132 |
| FKBP3 | FKBP-25 | PPIase | FK506 binding protein 3, 25kDa | Reconstituted Complex | BioGRID | 12920132 |
| GATAD2B | FLJ37346 | KIAA1150 | MGC138257 | MGC138285 | P66beta | RP11-216N14.6 | GATA zinc finger domain containing 2B | Affinity Capture-Western | BioGRID | 11756549 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | Affinity Capture-MS Affinity Capture-Western Co-purification | BioGRID | 10444591 |11171972 |12374985 |12920132 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | Affinity Capture-MS Affinity Capture-Western Co-purification | BioGRID | 10444591 |12493763 |12920132 |
| MBD2 | DKFZp586O0821 | DMTase | NY-CO-41 | methyl-CpG binding domain protein 2 | Reconstituted Complex | BioGRID | 10444591 |
| MBD3 | - | methyl-CpG binding domain protein 3 | MBD3 interacts with MTA2. | BIND | 12124384 |
| MBD3 | - | methyl-CpG binding domain protein 3 | - | HPRD,BioGRID | 12124384 |
| MBD3L1 | MBD3L | MGC138263 | MGC138269 | methyl-CpG binding domain protein 3-like 1 | Affinity Capture-Western | BioGRID | 15456747 |
| MTA1 | - | metastasis associated 1 | Affinity Capture-MS | BioGRID | 12920132 |
| MTA2 | DKFZp686F2281 | MTA1L1 | PID | metastasis associated 1 family, member 2 | Affinity Capture-MS | BioGRID | 12920132 |
| RBBP4 | NURF55 | RBAP48 | retinoblastoma binding protein 4 | Affinity Capture-Western Co-purification | BioGRID | 10444591 |12920132 |
| RBBP7 | MGC138867 | MGC138868 | RbAp46 | retinoblastoma binding protein 7 | Affinity Capture-Western Co-purification | BioGRID | 10444591 |12920132 |
| SAP30 | - | Sin3A-associated protein, 30kDa | Co-purification | BioGRID | 10444591 |
| SATB1 | - | SATB homeobox 1 | - | HPRD,BioGRID | 12374985 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | Affinity Capture-MS | BioGRID | 12920132 |
| SPEN | KIAA0929 | MINT | RP1-134O19.1 | SHARP | spen homolog, transcriptional regulator (Drosophila) | - | HPRD | 11331609 |
| TOP2B | TOPIIB | top2beta | topoisomerase (DNA) II beta 180kDa | Affinity Capture-Western | BioGRID | 11062478 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | Reconstituted Complex | BioGRID | 12920132 |
| TP53 | FLJ92943 | LFS1 | TRP53 | p53 | tumor protein p53 | PID interacts with p53. | BIND | 11099047 |
| YY1 | DELTA | INO80S | NF-E1 | UCRBP | YIN-YANG-1 | YY1 transcription factor | Reconstituted Complex | BioGRID | 12920132 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID HDAC CLASSI PATHWAY | 66 | 50 | All SZGR 2.0 genes in this pathway |
| WATANABE COLON CANCER MSI VS MSS UP | 29 | 18 | All SZGR 2.0 genes in this pathway |
| CAMPS COLON CANCER COPY NUMBER DN | 74 | 34 | All SZGR 2.0 genes in this pathway |