Gene Page: CD8A
Summary ?
| GeneID | 925 |
| Symbol | CD8A |
| Synonyms | CD8|Leu2|MAL|p32 |
| Description | CD8a molecule |
| Reference | MIM:186910|HGNC:HGNC:1706|Ensembl:ENSG00000153563|HPRD:01737|Vega:OTTHUMG00000130265 |
| Gene type | protein-coding |
| Map location | 2p12 |
| Pascal p-value | 0.004 |
| Fetal beta | -0.25 |
| DMG | 2 (# studies) |
| eGene | Cerebellum Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 3 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 3 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0004 | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03318654 | 2 | 87034200 | CD8A | 2.408E-4 | 0.45 | 0.037 | DMG:Wockner_2014 |
| cg02870945 | 2 | 86565124 | CD8A | 6.249E-4 | 11.142 | DMG:vanEijk_2014 | |
| cg02870945 | 2 | 86565124 | CD8A | 0.003 | 10.047 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
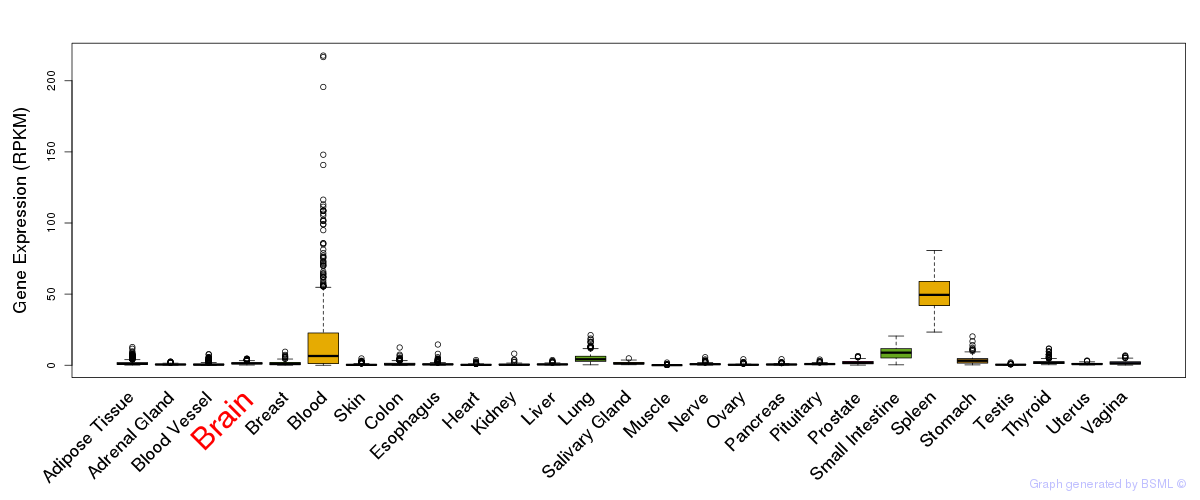
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MMP25 | 0.61 | 0.56 |
| KCTD11 | 0.57 | 0.56 |
| DPEP1 | 0.57 | 0.54 |
| NT5DC2 | 0.57 | 0.54 |
| AC006273.1 | 0.56 | 0.51 |
| ISYNA1 | 0.56 | 0.55 |
| COMTD1 | 0.56 | 0.47 |
| PPDPF | 0.56 | 0.46 |
| PVRL2 | 0.56 | 0.51 |
| RAC3 | 0.56 | 0.46 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ALDOC | -0.43 | -0.45 |
| RGN | -0.42 | -0.47 |
| FBXO2 | -0.42 | -0.44 |
| OMG | -0.41 | -0.48 |
| C5orf53 | -0.41 | -0.48 |
| CSRP1 | -0.41 | -0.45 |
| CCNI2 | -0.40 | -0.44 |
| SPARCL1 | -0.40 | -0.44 |
| CA2 | -0.39 | -0.47 |
| ANXA11 | -0.39 | -0.44 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 2470098 |2493728 | |
| GO:0042288 | MHC class I protein binding | NAS | 11131152 | |
| GO:0015026 | coreceptor activity | NAS | 11131152 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0002456 | T cell mediated immunity | IEA | - | |
| GO:0007169 | transmembrane receptor protein tyrosine kinase signaling pathway | NAS | 9830036 | |
| GO:0007166 | cell surface receptor linked signal transduction | IEA | - | |
| GO:0019882 | antigen processing and presentation | NAS | 2496167 | |
| GO:0006955 | immune response | NAS | 11131152 | |
| GO:0042110 | T cell activation | NAS | 9830036 | |
| GO:0050850 | positive regulation of calcium-mediated signaling | IEA | - | |
| GO:0045065 | cytotoxic T cell differentiation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0042101 | T cell receptor complex | NAS | Synap (GO term level: 8) | 11131152 |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0009897 | external side of plasma membrane | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | NAS | 2496167 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CD3D | CD3-DELTA | T3D | CD3d molecule, delta (CD3-TCR complex) | - | HPRD,BioGRID | 12215456 |
| CD59 | 16.3A5 | 1F5 | EJ16 | EJ30 | EL32 | FLJ38134 | FLJ92039 | G344 | HRF-20 | HRF20 | MAC-IP | MACIF | MEM43 | MGC2354 | MIC11 | MIN1 | MIN2 | MIN3 | MIRL | MSK21 | p18-20 | CD59 molecule, complement regulatory protein | Reconstituted Complex | BioGRID | 1377690 |
| CD8A | CD8 | Leu2 | MAL | p32 | CD8a molecule | Co-crystal Structure Co-localization | BioGRID | 8127870 |9550407 |
| CD8A | CD8 | Leu2 | MAL | p32 | CD8a molecule | - | HPRD | 8127870 |10809759 |
| CD8B | CD8B1 | LYT3 | Leu2 | Ly3 | MGC119115 | CD8b molecule | - | HPRD,BioGRID | 3145195 |9550407 |10623829|10809759 |
| HLA-A | HLAA | major histocompatibility complex, class I, A | - | HPRD,BioGRID | 9177355 |10809759|10809759 |
| HLA-B | AS | HLA-B-7301 | HLA-B73 | HLAB | HLAC | SPDA1 | major histocompatibility complex, class I, B | - | HPRD,BioGRID | 9331948 |10809759|10809759 |
| HLA-C | D6S204 | FLJ27082 | HLA-Cw | HLA-Cw12 | HLA-JY3 | HLC-C | PSORS1 | major histocompatibility complex, class I, C | - | HPRD,BioGRID | 10809759 |
| HLA-E | DKFZp686P19218 | EA1.2 | EA2.1 | HLA-6.2 | MHC | QA1 | major histocompatibility complex, class I, E | - | HPRD | 10809759 |
| HLA-G | MHC-G | major histocompatibility complex, class I, G | - | HPRD | 1908512 |10843658|10809759 |
| HLA-G | MHC-G | major histocompatibility complex, class I, G | in vivo Reconstituted Complex | BioGRID | 1908512 |10809759 |10843658 |
| LAT | LAT1 | pp36 | linker for activation of T cells | - | HPRD | 10562325 |
| LGALS1 | DKFZp686E23103 | GAL1 | GBP | lectin, galactoside-binding, soluble, 1 | - | HPRD,BioGRID | 10490978 |
| PTPRC | B220 | CD45 | CD45R | GP180 | LCA | LY5 | T200 | protein tyrosine phosphatase, receptor type, C | - | HPRD,BioGRID | 1834739 |
| SFRS12 | DKFZp564B176 | MGC133045 | SRrp508 | SRrp86 | splicing factor, arginine/serine-rich 12 | - | HPRD | 14559993 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CELL ADHESION MOLECULES CAMS | 134 | 93 | All SZGR 2.0 genes in this pathway |
| KEGG ANTIGEN PROCESSING AND PRESENTATION | 89 | 65 | All SZGR 2.0 genes in this pathway |
| KEGG HEMATOPOIETIC CELL LINEAGE | 88 | 60 | All SZGR 2.0 genes in this pathway |
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| KEGG PRIMARY IMMUNODEFICIENCY | 35 | 28 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL17 PATHWAY | 17 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA STEM PATHWAY | 15 | 13 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TCYTOTOXIC PATHWAY | 14 | 11 | All SZGR 2.0 genes in this pathway |
| PID IL12 2PATHWAY | 63 | 54 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR PATHWAY | 53 | 42 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR DOWNSTREAM PATHWAY | 65 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | 70 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA UP | 177 | 110 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL DN | 226 | 132 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 4 5WK DN | 196 | 131 | All SZGR 2.0 genes in this pathway |
| MCBRYAN PUBERTAL BREAST 5 6WK UP | 116 | 68 | All SZGR 2.0 genes in this pathway |
| TANAKA METHYLATED IN ESOPHAGEAL CARCINOMA | 103 | 58 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER CLUSTER 1 | 44 | 23 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM5 | 94 | 59 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK UP | 87 | 58 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| HOWLIN CITED1 TARGETS 1 UP | 35 | 25 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA RADIATION | 81 | 59 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE UP | 226 | 164 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| LEE DOUBLE POLAR THYMOCYTE | 27 | 17 | All SZGR 2.0 genes in this pathway |
| FINAK BREAST CANCER SDPP SIGNATURE | 26 | 11 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| CHIARETTI T ALL RELAPSE PROGNOSIS | 19 | 15 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-186 | 151 | 157 | 1A | hsa-miR-186 | CAAAGAAUUCUCCUUUUGGGCUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.