| ONKEN UVEAL MELANOMA DN
| 526 | 357 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA PROGENITOR UP
| 39 | 25 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP
| 473 | 314 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE UP
| 485 | 293 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL UP
| 233 | 161 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA UP
| 544 | 308 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS UP
| 214 | 155 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN
| 508 | 354 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE EARLY
| 66 | 48 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO IONIZING RADIATION
| 149 | 101 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP
| 1037 | 673 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN
| 428 | 306 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215
| 893 | 528 | All SZGR 2.0 genes in this pathway |
| ZHANG RESPONSE TO IKK INHIBITOR AND TNF UP
| 223 | 140 | All SZGR 2.0 genes in this pathway |
| MORI LARGE PRE BII LYMPHOCYTE DN
| 58 | 39 | All SZGR 2.0 genes in this pathway |
| MORI IMMATURE B LYMPHOCYTE UP
| 53 | 35 | All SZGR 2.0 genes in this pathway |
| MORI MATURE B LYMPHOCYTE UP
| 90 | 62 | All SZGR 2.0 genes in this pathway |
| MORI PLASMA CELL DN
| 33 | 20 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 6P24 P22 AMPLICON
| 21 | 17 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN
| 464 | 276 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 SIGNALING VIA CTNNB1
| 83 | 58 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP
| 266 | 171 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS B LYMPHOCYTE DN
| 38 | 25 | All SZGR 2.0 genes in this pathway |
| WIELAND UP BY HBV INFECTION
| 101 | 66 | All SZGR 2.0 genes in this pathway |
| GALINDO IMMUNE RESPONSE TO ENTEROTOXIN
| 85 | 67 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION UP
| 114 | 84 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING UP
| 101 | 76 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION
| 405 | 264 | All SZGR 2.0 genes in this pathway |
| JAZAERI BREAST CANCER BRCA1 VS BRCA2 DN
| 43 | 31 | All SZGR 2.0 genes in this pathway |
| KAYO AGING MUSCLE UP
| 244 | 165 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 18HR UP
| 178 | 111 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 24HR UP
| 148 | 96 | All SZGR 2.0 genes in this pathway |
| BILD E2F3 ONCOGENIC SIGNATURE
| 246 | 153 | All SZGR 2.0 genes in this pathway |
| LEIN MIDBRAIN MARKERS
| 82 | 55 | All SZGR 2.0 genes in this pathway |
| LEIN PONS MARKERS
| 89 | 59 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P4
| 100 | 62 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN
| 409 | 268 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP
| 299 | 167 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN
| 270 | 181 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP
| 425 | 298 | All SZGR 2.0 genes in this pathway |
| VILIMAS NOTCH1 TARGETS UP
| 52 | 41 | All SZGR 2.0 genes in this pathway |
| LINDSTEDT DENDRITIC CELL MATURATION B
| 53 | 36 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP
| 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN
| 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN
| 245 | 150 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS KERATINOCYTE UP
| 91 | 63 | All SZGR 2.0 genes in this pathway |
| HINATA NFKB TARGETS FIBROBLAST UP
| 84 | 60 | All SZGR 2.0 genes in this pathway |
| HOFFMANN SMALL PRE BII TO IMMATURE B LYMPHOCYTE UP
| 70 | 49 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK
| 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED
| 536 | 296 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3
| 1069 | 729 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS
| 535 | 325 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 12HR DN
| 33 | 25 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 24HR DN
| 43 | 32 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 32HR DN
| 39 | 28 | All SZGR 2.0 genes in this pathway |
| DORN ADENOVIRUS INFECTION 48HR DN
| 40 | 29 | All SZGR 2.0 genes in this pathway |
| TIAN TNF SIGNALING VIA NFKB
| 28 | 21 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 9
| 76 | 45 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS UP
| 217 | 131 | All SZGR 2.0 genes in this pathway |
| WIERENGA STAT5A TARGETS GROUP2
| 60 | 38 | All SZGR 2.0 genes in this pathway |
| PHONG TNF TARGETS UP
| 63 | 43 | All SZGR 2.0 genes in this pathway |
| PHONG TNF RESPONSE NOT VIA P38
| 337 | 236 | All SZGR 2.0 genes in this pathway |
| BOSCO ALLERGEN INDUCED TH2 ASSOCIATED MODULE
| 151 | 86 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 2HR UP
| 53 | 33 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY
| 1725 | 838 | All SZGR 2.0 genes in this pathway |
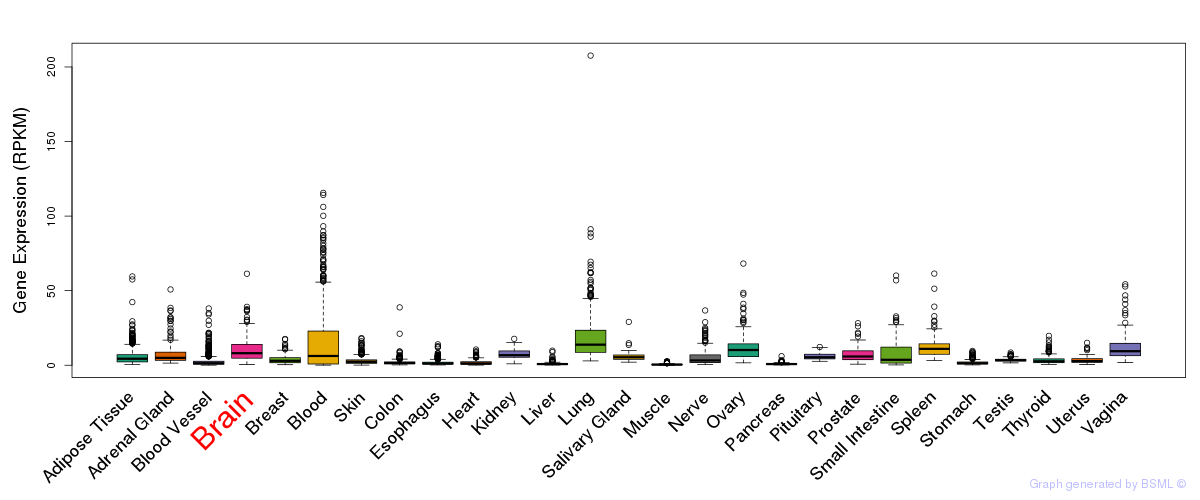
 eQTL annotation
eQTL annotation