Gene Page: DEPDC5
Summary ?
| GeneID | 9681 |
| Symbol | DEPDC5 |
| Synonyms | DEP.5|FFEVF |
| Description | DEP domain containing 5 |
| Reference | MIM:614191|HGNC:HGNC:18423|Ensembl:ENSG00000100150|HPRD:16795|Vega:OTTHUMG00000030926 |
| Gene type | protein-coding |
| Map location | 22q12.3 |
| Pascal p-value | 0.338 |
| Sherlock p-value | 0.455 |
| Fetal beta | -0.067 |
| DMG | 2 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg25798026 | 22 | 32149910 | DEPDC5 | 1.664E-4 | -0.24 | 0.033 | DMG:Wockner_2014 |
| cg03550506 | 22 | 32149733 | DEPDC5 | 1.37E-9 | -0.015 | 1.37E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
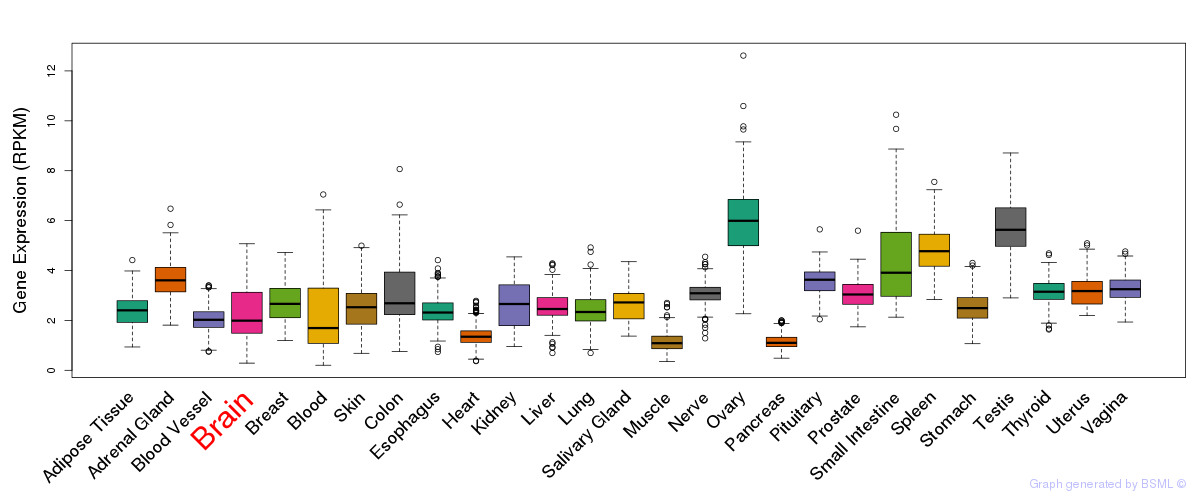
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ATG9A | 0.93 | 0.92 |
| NFE2L1 | 0.91 | 0.91 |
| MAPK8IP3 | 0.91 | 0.89 |
| ZER1 | 0.91 | 0.89 |
| AGPAT3 | 0.90 | 0.90 |
| DAGLA | 0.90 | 0.91 |
| AMIGO1 | 0.90 | 0.90 |
| DENND4B | 0.90 | 0.88 |
| TECPR2 | 0.89 | 0.89 |
| KIAA0513 | 0.88 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GNG11 | -0.57 | -0.59 |
| AF347015.21 | -0.55 | -0.50 |
| DBI | -0.54 | -0.60 |
| C1orf54 | -0.53 | -0.57 |
| SYCP3 | -0.53 | -0.61 |
| IL32 | -0.52 | -0.53 |
| AL050337.1 | -0.51 | -0.58 |
| C1orf61 | -0.49 | -0.51 |
| CLEC2B | -0.48 | -0.50 |
| FAM159B | -0.47 | -0.66 |
Section III. Gene Ontology annotation
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0007242 | intracellular signaling cascade | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE UP | 134 | 93 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| LIN APC TARGETS | 77 | 55 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES DN | 210 | 141 | All SZGR 2.0 genes in this pathway |