Gene Page: SEMA3E
Summary ?
| GeneID | 9723 |
| Symbol | SEMA3E |
| Synonyms | M-SEMAH|M-SemaK|SEMAH|coll-5 |
| Description | semaphorin 3E |
| Reference | MIM:608166|HGNC:HGNC:10727|Ensembl:ENSG00000170381|HPRD:11545|Vega:OTTHUMG00000154693 |
| Gene type | protein-coding |
| Map location | 7q21.11 |
| Pascal p-value | 0.481 |
| Sherlock p-value | 0.667 |
| Fetal beta | 1.032 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SEMA3E | chr7 | 83036424 | G | T | NM_001178129 NM_012431 | . . | silent silent | Schizophrenia | DNM:Gulsuner_2013 |
Section II. Transcriptome annotation
General gene expression (GTEx)
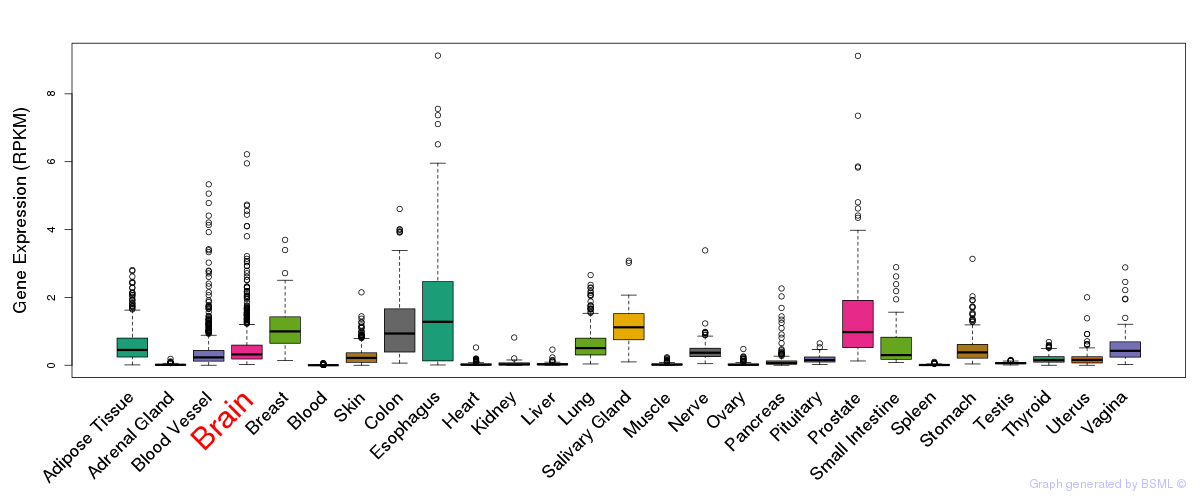
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TRIM23 | 0.96 | 0.96 |
| OPA1 | 0.95 | 0.95 |
| CLTC | 0.95 | 0.94 |
| PPM1A | 0.93 | 0.94 |
| DMXL2 | 0.93 | 0.93 |
| PRKACB | 0.93 | 0.94 |
| SRPK2 | 0.93 | 0.96 |
| WDR7 | 0.93 | 0.94 |
| SERINC1 | 0.92 | 0.93 |
| AKAP11 | 0.92 | 0.95 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RAB34 | -0.63 | -0.71 |
| EIF4EBP3 | -0.59 | -0.65 |
| GSDMD | -0.58 | -0.61 |
| EFEMP2 | -0.58 | -0.63 |
| FXYD1 | -0.57 | -0.59 |
| C1orf61 | -0.57 | -0.74 |
| GATSL3 | -0.57 | -0.62 |
| HIGD1B | -0.57 | -0.58 |
| AF347015.21 | -0.56 | -0.54 |
| C19orf36 | -0.56 | -0.63 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007399 | nervous system development | IEA | neurite (GO term level: 5) | - |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005615 | extracellular space | TAS | 9515811 | |
| GO:0016020 | membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG AXON GUIDANCE | 129 | 103 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME OTHER SEMAPHORIN INTERACTIONS | 15 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMAPHORIN INTERACTIONS | 68 | 53 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| LEE TARGETS OF PTCH1 AND SUFU DN | 83 | 69 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS DN | 31 | 23 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS DN | 135 | 88 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN DN | 353 | 226 | All SZGR 2.0 genes in this pathway |
| OSADA ASCL1 TARGETS UP | 46 | 30 | All SZGR 2.0 genes in this pathway |
| LABBE TGFB1 TARGETS DN | 108 | 64 | All SZGR 2.0 genes in this pathway |
| LABBE TARGETS OF TGFB1 AND WNT3A DN | 108 | 68 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| WONG ADULT TISSUE STEM MODULE | 721 | 492 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 3 | 15 | 7 | All SZGR 2.0 genes in this pathway |
| NOUSHMEHR GBM SILENCED BY METHYLATION | 50 | 32 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS NOT VIA AKT1 DN | 88 | 61 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 DN | 82 | 51 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| PASINI SUZ12 TARGETS DN | 315 | 215 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
| NABA ECM AFFILIATED | 171 | 89 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |