Gene Page: DHX38
Summary ?
| GeneID | 9785 |
| Symbol | DHX38 |
| Synonyms | DDX38|PRP16|PRPF16 |
| Description | DEAH-box helicase 38 |
| Reference | MIM:605584|HGNC:HGNC:17211|Ensembl:ENSG00000140829|HPRD:05720|Vega:OTTHUMG00000137596 |
| Gene type | protein-coding |
| Map location | 16q22 |
| Pascal p-value | 0.543 |
| Sherlock p-value | 0.046 |
| Fetal beta | 0.773 |
| eGene | Myers' cis & trans Meta |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1026400 | chr21 | 43587823 | DHX38 | 9785 | 0.18 | trans |
Section II. Transcriptome annotation
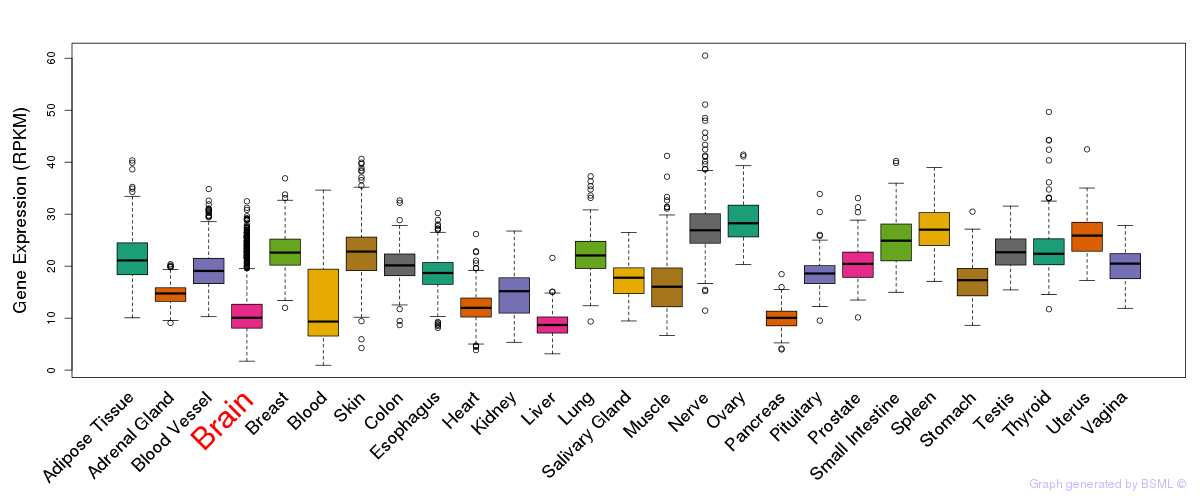
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC020663.1 | 0.91 | 0.90 |
| NOLC1 | 0.91 | 0.90 |
| MSL1 | 0.91 | 0.90 |
| VPS26B | 0.90 | 0.90 |
| C6orf168 | 0.90 | 0.89 |
| KIF3C | 0.89 | 0.91 |
| UBP1 | 0.89 | 0.89 |
| ATCAY | 0.89 | 0.91 |
| FBXW2 | 0.89 | 0.89 |
| XPO7 | 0.89 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.81 | -0.80 |
| MT-CO2 | -0.80 | -0.79 |
| AF347015.21 | -0.79 | -0.82 |
| AF347015.8 | -0.78 | -0.78 |
| AF347015.27 | -0.77 | -0.79 |
| MT-CYB | -0.77 | -0.76 |
| FXYD1 | -0.77 | -0.77 |
| AF347015.33 | -0.76 | -0.74 |
| HIGD1B | -0.75 | -0.75 |
| AF347015.2 | -0.75 | -0.73 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG SPLICEOSOME | 128 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME PROCESSING OF CAPPED INTRON CONTAINING PRE MRNA | 140 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | 54 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME RNA POL II TRANSCRIPTION | 105 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA PROCESSING | 161 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA SPLICING | 111 | 58 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTION | 210 | 127 | All SZGR 2.0 genes in this pathway |
| REACTOME MRNA 3 END PROCESSING | 35 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME CLEAVAGE OF GROWING TRANSCRIPT IN THE TERMINATION REGION | 44 | 22 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| LEE AGING CEREBELLUM DN | 86 | 66 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C0 | 107 | 72 | All SZGR 2.0 genes in this pathway |
| KAYO CALORIE RESTRICTION MUSCLE UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| JIANG TIP30 TARGETS DN | 24 | 14 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C UP | 47 | 29 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |