Gene Page: PHYHIP
Summary ?
| GeneID | 9796 |
| Symbol | PHYHIP |
| Synonyms | DYRK1AP3|PAHX-AP|PAHXAP1 |
| Description | phytanoyl-CoA 2-hydroxylase interacting protein |
| Reference | MIM:608511|HGNC:HGNC:16865|Ensembl:ENSG00000168490|HPRD:16345|Vega:OTTHUMG00000163776 |
| Gene type | protein-coding |
| Map location | 8p21.3 |
| Pascal p-value | 0.38 |
| Sherlock p-value | 0.154 |
| DEG p-value | DEG:Maycox_2009:CC_BA10_fold_change=-1.11:CC_BA10_disease_P=0.0217:HBB_BA9_fold_change=-1.24:HBB_BA9_disease_P=0.0007 |
| Fetal beta | -3.125 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Cerebellum Myers' cis & trans |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Maycox_2009 | Microarray to determine the expression of over 30000 mRNA transcripts in post-mortem tissue | We included 51 genes whose expression changes are common between two schizophrenia cohorts. | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.031 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.03086 | |
| GSMA_IIE | Genome scan meta-analysis (European-ancestry samples) | Psr: 0.00057 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg22102057 | 8 | 22084298 | PHYHIP | 7.25E-5 | 0.742 | 0.025 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4854166 | chr2 | 3297005 | PHYHIP | 9796 | 0.17 | trans |
Section II. Transcriptome annotation
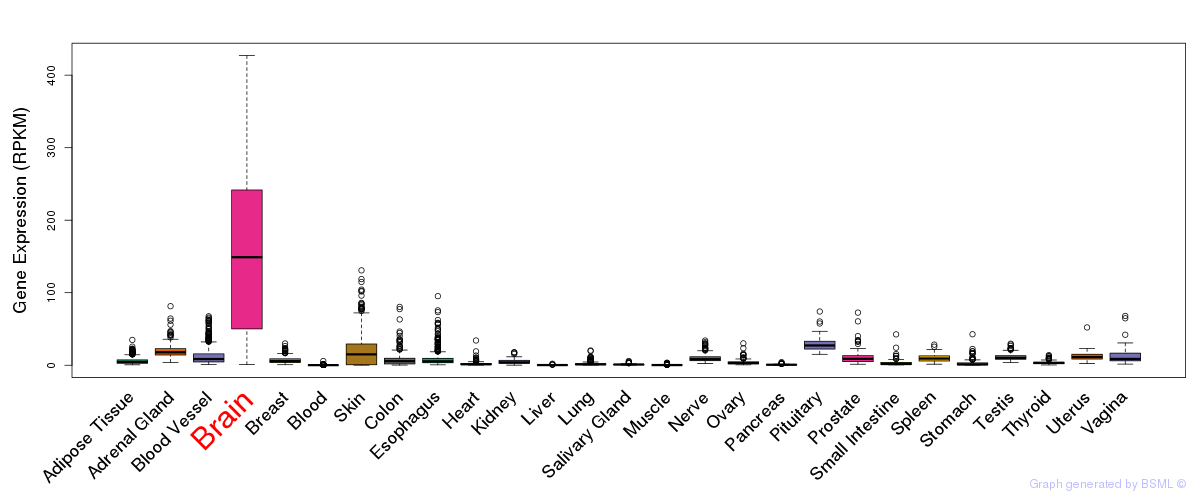
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C1orf92 | 0.95 | 0.50 |
| C3orf25 | 0.95 | 0.59 |
| CCDC114 | 0.95 | 0.50 |
| WDR93 | 0.94 | 0.56 |
| WDR63 | 0.93 | 0.43 |
| CCDC108 | 0.93 | 0.64 |
| RIBC1 | 0.93 | 0.59 |
| CCDC11 | 0.93 | 0.54 |
| CCDC135 | 0.93 | 0.53 |
| CCDC19 | 0.93 | 0.42 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CIDEA | -0.20 | -0.24 |
| NUAK1 | -0.19 | -0.19 |
| RGS6 | -0.18 | -0.24 |
| IER5L | -0.18 | -0.18 |
| ANKRD56 | -0.17 | -0.13 |
| FBXW7 | -0.17 | -0.15 |
| EMX1 | -0.17 | -0.22 |
| KCNV1 | -0.17 | -0.10 |
| LNX1 | -0.17 | -0.13 |
| SLC26A4 | -0.16 | -0.14 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BAI1 | FLJ41988 | brain-specific angiogenesis inhibitor 1 | - | HPRD | 11245925 |
| C14orf1 | ERG28 | NET51 | chromosome 14 open reading frame 1 | Two-hybrid | BioGRID | 16169070 |
| C1orf156 | AsTP2 | MGC9084 | RP1-117P20.4 | chromosome 1 open reading frame 156 | Two-hybrid | BioGRID | 16169070 |
| COPS6 | CSN6 | MOV34-34KD | COP9 constitutive photomorphogenic homolog subunit 6 (Arabidopsis) | Two-hybrid | BioGRID | 16169070 |
| DYRK1A | DYRK | DYRK1 | HP86 | MNB | MNBH | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A | DYRK1A interacts with PAHX-AP1. | BIND | 15694837 |
| DYRK1A | DYRK | DYRK1 | HP86 | MNB | MNBH | dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A | - | HPRD | 15694837 |
| EEF1A1 | CCS-3 | CCS3 | EEF-1 | EEF1A | EF-Tu | EF1A | FLJ25721 | GRAF-1EF | HNGC:16303 | LENG7 | MGC102687 | MGC131894 | MGC16224 | PTI1 | eEF1A-1 | eukaryotic translation elongation factor 1 alpha 1 | Two-hybrid | BioGRID | 16169070 |
| FAM131A | C3orf40 | FLAT715 | MGC21688 | PRO1378 | family with sequence similarity 131, member A | Two-hybrid | BioGRID | 16169070 |
| HDAC11 | FLJ22237 | histone deacetylase 11 | Two-hybrid | BioGRID | 16169070 |
| MAGED4B | MGC3210 | MGC88639 | melanoma antigen family D, 4B | Two-hybrid | BioGRID | 16169070 |
| MED8 | ARC32 | MGC17544 | MGC19641 | mediator complex subunit 8 | Two-hybrid | BioGRID | 16169070 |
| NDRG1 | CAP43 | CMT4D | DRG1 | GC4 | HMSNL | NDR1 | NMSL | PROXY1 | RIT42 | RTP | TARG1 | TDD5 | N-myc downstream regulated 1 | Two-hybrid | BioGRID | 16169070 |
| NDUFV3 | CI-9KD | NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa | Two-hybrid | BioGRID | 16169070 |
| PHYH | LN1 | LNAP1 | PAHX | PHYH1 | RD | phytanoyl-CoA 2-hydroxylase | - | HPRD | 10686344 |
| PNPLA2 | 1110001C14Rik | ATGL | DESNUTRIN | DKFZp667M109 | FP17548 | PEDF-R | TTS-2.2 | TTS2 | patatin-like phospholipase domain containing 2 | Two-hybrid | BioGRID | 16169070 |
| PPIE | CYP-33 | MGC111222 | MGC3736 | peptidylprolyl isomerase E (cyclophilin E) | Two-hybrid | BioGRID | 16169070 |
| PRMT5 | HRMT1L5 | IBP72 | JBP1 | SKB1 | SKB1Hs | protein arginine methyltransferase 5 | Two-hybrid | BioGRID | 16169070 |
| S100A13 | - | S100 calcium binding protein A13 | Two-hybrid | BioGRID | 16169070 |
| SMARCC2 | BAF170 | CRACC2 | Rsc8 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily c, member 2 | Two-hybrid | BioGRID | 16169070 |
| TTR | HsT2651 | PALB | TBPA | transthyretin | Two-hybrid | BioGRID | 16169070 |
| WDR89 | C14orf150 | MGC9907 | MSTP050 | WD repeat domain 89 | Two-hybrid | BioGRID | 16169070 |
| ZZEF1 | FLJ10821 | FLJ23789 | FLJ45574 | KIAA0399 | MGC166929 | ZZZ4 | zinc finger, ZZ-type with EF-hand domain 1 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| VANHARANTA UTERINE FIBROID DN | 67 | 45 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF ICP WITH H3K27ME3 | 206 | 108 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS ICP WITH H3K4ME3 AND H327ME3 | 126 | 83 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 AND H3K27ME3 | 137 | 85 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS DN | 242 | 146 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-125/351 | 446 | 452 | m8 | hsa-miR-125bbrain | UCCCUGAGACCCUAACUUGUGA |
| hsa-miR-125abrain | UCCCUGAGACCCUUUAACCUGUG | ||||
| miR-140 | 1612 | 1618 | m8 | hsa-miR-140brain | AGUGGUUUUACCCUAUGGUAG |
| miR-185 | 82 | 88 | m8 | hsa-miR-185brain | UGGAGAGAAAGGCAGUUC |
| miR-299-5p | 1611 | 1617 | 1A | hsa-miR-299-5p | UGGUUUACCGUCCCACAUACAU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.