Gene Page: ARHGEF11
Summary ?
| GeneID | 9826 |
| Symbol | ARHGEF11 |
| Synonyms | GTRAP48|PDZ-RHOGEF |
| Description | Rho guanine nucleotide exchange factor 11 |
| Reference | MIM:605708|HGNC:HGNC:14580|Ensembl:ENSG00000132694|HPRD:09302|Vega:OTTHUMG00000041292 |
| Gene type | protein-coding |
| Map location | 1q21 |
| Sherlock p-value | 0.519 |
| Fetal beta | -0.636 |
| eGene | Myers' cis & trans |
| Support | CompositeSet Darnell FMRP targets Ascano FMRP targets Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.0235 | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.00814 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ARHGEF11 | chr1 | 156911204 | G | A | NM_014784 NM_198236 | . . | silent silent | Schizophrenia | DNM:Fromer_2014 | ||
| ARHGEF11 | chr1 | 156918274 | G | A | NM_014784 NM_198236 | p.608R>C p.648R>C | missense missense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6128541 | chr20 | 57917688 | ARHGEF11 | 9826 | 0.07 | trans |
Section II. Transcriptome annotation
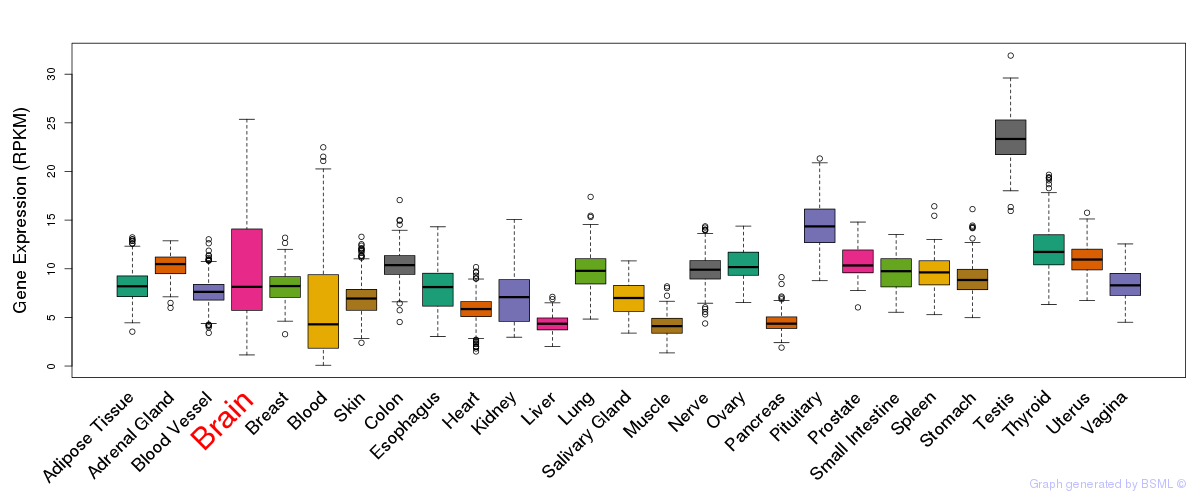
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SNAP47 | 0.86 | 0.81 |
| TMEM55B | 0.85 | 0.80 |
| TOMM40L | 0.84 | 0.79 |
| ATG4B | 0.84 | 0.80 |
| ARFIP2 | 0.84 | 0.74 |
| UBL4A | 0.83 | 0.78 |
| ST3GAL3 | 0.83 | 0.81 |
| C7orf20 | 0.82 | 0.78 |
| TBRG4 | 0.82 | 0.76 |
| GSK3A | 0.82 | 0.77 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.71 | -0.64 |
| AF347015.8 | -0.70 | -0.60 |
| MT-CO2 | -0.68 | -0.58 |
| AF347015.18 | -0.67 | -0.63 |
| AF347015.26 | -0.67 | -0.58 |
| MT-CYB | -0.67 | -0.59 |
| AF347015.2 | -0.67 | -0.57 |
| AF347015.31 | -0.67 | -0.58 |
| MT-ATP8 | -0.66 | -0.64 |
| AF347015.27 | -0.66 | -0.61 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005089 | Rho guanyl-nucleotide exchange factor activity | IEA | - | |
| GO:0005089 | Rho guanyl-nucleotide exchange factor activity | TAS | 10026210 | |
| GO:0005085 | guanyl-nucleotide exchange factor activity | IEA | - | |
| GO:0001664 | G-protein-coupled receptor binding | IDA | 15755723 | |
| GO:0004871 | signal transducer activity | IEA | - | |
| GO:0005096 | GTPase activator activity | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 10026210 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001558 | regulation of cell growth | NAS | 10026210 | |
| GO:0000910 | cytokinesis | NAS | 10026210 | |
| GO:0007186 | G-protein coupled receptor protein signaling pathway | TAS | 10026210 | |
| GO:0030010 | establishment of cell polarity | NAS | 10026210 | |
| GO:0006941 | striated muscle contraction | NAS | 10026210 | |
| GO:0006928 | cell motion | NAS | 10026210 | |
| GO:0030036 | actin cytoskeleton organization | NAS | 10026210 | |
| GO:0035023 | regulation of Rho protein signal transduction | IEA | - | |
| GO:0045893 | positive regulation of transcription, DNA-dependent | IDA | 10026210 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005622 | intracellular | IC | 10026210 | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005737 | cytoplasm | IDA | 15755723 | |
| GO:0016020 | membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG VASCULAR SMOOTH MUSCLE CONTRACTION | 115 | 81 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MYOSIN PATHWAY | 31 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RHO PATHWAY | 32 | 23 | All SZGR 2.0 genes in this pathway |
| BIOCARTA PAR1 PATHWAY | 37 | 28 | All SZGR 2.0 genes in this pathway |
| SIG CHEMOTAXIS | 45 | 37 | All SZGR 2.0 genes in this pathway |
| ST GA13 PATHWAY | 37 | 32 | All SZGR 2.0 genes in this pathway |
| PID RHOA REG PATHWAY | 46 | 30 | All SZGR 2.0 genes in this pathway |
| PID FAK PATHWAY | 59 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY RHO GTPASES | 113 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME NRAGE SIGNALS DEATH THROUGH JNK | 43 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | 60 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | 81 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME AXON GUIDANCE | 251 | 188 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA1213 SIGNALLING EVENTS | 74 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMA4D IN SEMAPHORIN SIGNALING | 32 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMAPHORIN INTERACTIONS | 68 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | 27 | 21 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 UP | 341 | 197 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 LCP WITH H3K4ME3 | 162 | 80 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF LCP WITH H3K4ME3 | 128 | 68 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS LCP WITH H3K4ME3 | 174 | 100 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES LCP WITH H3K4ME3 | 142 | 80 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC LCP WITH H3K4ME3 | 58 | 34 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-128 | 995 | 1001 | m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-132/212 | 1070 | 1076 | m8 | hsa-miR-212SZ | UAACAGUCUCCAGUCACGGCC |
| hsa-miR-132brain | UAACAGUCUACAGCCAUGGUCG | ||||
| miR-17-5p/20/93.mr/106/519.d | 374 | 380 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.