Gene Page: SGSM2
Summary ?
| GeneID | 9905 |
| Symbol | SGSM2 |
| Synonyms | RUTBC1 |
| Description | small G protein signaling modulator 2 |
| Reference | MIM:611418|HGNC:HGNC:29026|Ensembl:ENSG00000141258|HPRD:15284|Vega:OTTHUMG00000177637 |
| Gene type | protein-coding |
| Map location | 17p13.3 |
| Pascal p-value | 1.838E-6 |
| Sherlock p-value | 0.171 |
| TADA p-value | 0.021 |
| Fetal beta | 0.016 |
| Support | CompositeSet Darnell FMRP targets Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| SGSM2 | chr17 | 2281220 | C | T | NM_001098509 NM_014853 | p.913R>C p.958R>C | missense missense | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
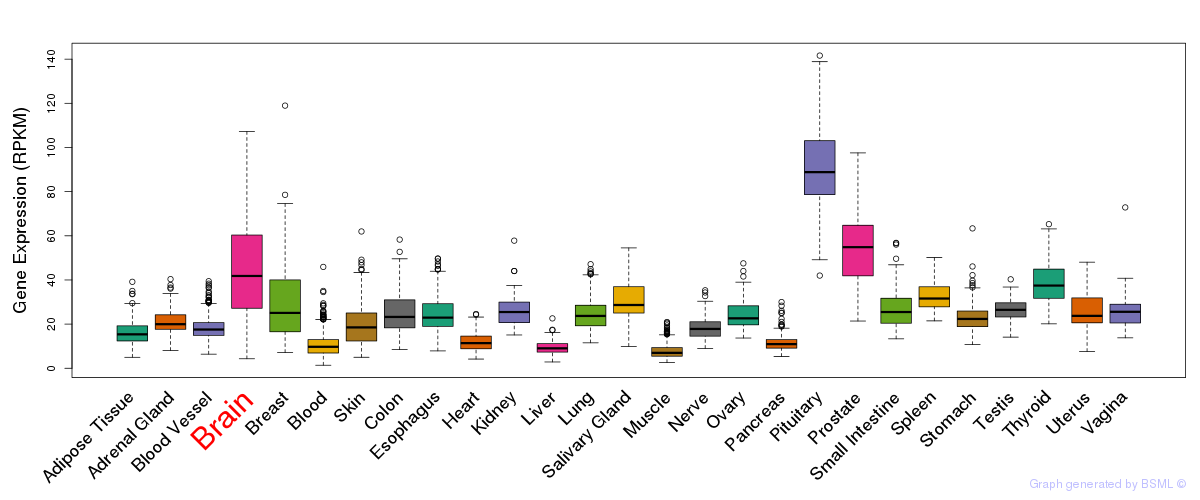
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AFP | FETA | HPAFP | alpha-fetoprotein | Two-hybrid | BioGRID | 16169070 |
| CA12 | CAXII | FLJ20151 | HsT18816 | carbonic anhydrase XII | Two-hybrid | BioGRID | 16169070 |
| CELSR3 | CDHF11 | EGFL1 | FMI1 | HFMI1 | MEGF2 | RESDA1 | cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) | Two-hybrid | BioGRID | 16169070 |
| CHD3 | Mi-2a | Mi2-ALPHA | ZFH | chromodomain helicase DNA binding protein 3 | Two-hybrid | BioGRID | 16169070 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | Two-hybrid | BioGRID | 16169070 |
| FNBP1L | C1orf39 | TOCA1 | formin binding protein 1-like | Two-hybrid | BioGRID | 16169070 |
| GDPD2 | FLJ20207 | GDE3 | OBDPF | glycerophosphodiester phosphodiesterase domain containing 2 | Two-hybrid | BioGRID | 16169070 |
| KHDRBS3 | Etle | SALP | SLM-2 | SLM2 | T-STAR | TSTAR | etoile | KH domain containing, RNA binding, signal transduction associated 3 | Two-hybrid | BioGRID | 16169070 |
| MPP1 | AAG12 | DXS552 | DXS552E | EMP55 | MRG1 | PEMP | membrane protein, palmitoylated 1, 55kDa | Two-hybrid | BioGRID | 16169070 |
| PNO1 | KHRBP1 | partner of NOB1 homolog (S. cerevisiae) | Two-hybrid | BioGRID | 16169070 |
| PTN | HARP | HBGF8 | HBNF | NEGF1 | pleiotrophin | Two-hybrid | BioGRID | 16169070 |
| RGMA | RGM | RGM domain family, member A | Two-hybrid | BioGRID | 16169070 |
| RPS25 | - | ribosomal protein S25 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE FIMA DN | 284 | 156 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| GARCIA TARGETS OF FLI1 AND DAX1 UP | 57 | 34 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5 | 482 | 296 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR DN | 546 | 362 | All SZGR 2.0 genes in this pathway |
| WINTER HYPOXIA METAGENE | 242 | 168 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A4 | 196 | 124 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |