Gene Page: ADAMTS6
Summary ?
| GeneID | 11174 |
| Symbol | ADAMTS6 |
| Synonyms | ADAM-TS 6|ADAM-TS6|ADAMTS-6 |
| Description | ADAM metallopeptidase with thrombospondin type 1 motif 6 |
| Reference | MIM:605008|HGNC:HGNC:222|Ensembl:ENSG00000049192|HPRD:05421|Vega:OTTHUMG00000074079 |
| Gene type | protein-coding |
| Map location | 5q12 |
| Pascal p-value | 0.007 |
| Fetal beta | 1.408 |
| eGene | Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWAScat | Genome-wide Association Studies | This data set includes 560 SNPs associated with schizophrenia. A total of 486 genes were mapped to these SNPs within 50kb. | |
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ADAMTS6 | chr5 | 64466464 | G | T | NM_197941 | p.1075T>N | missense | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10994209 | chr10 | 61877705 | ADAMTS6 | 11174 | 0.2 | trans |
Section II. Transcriptome annotation
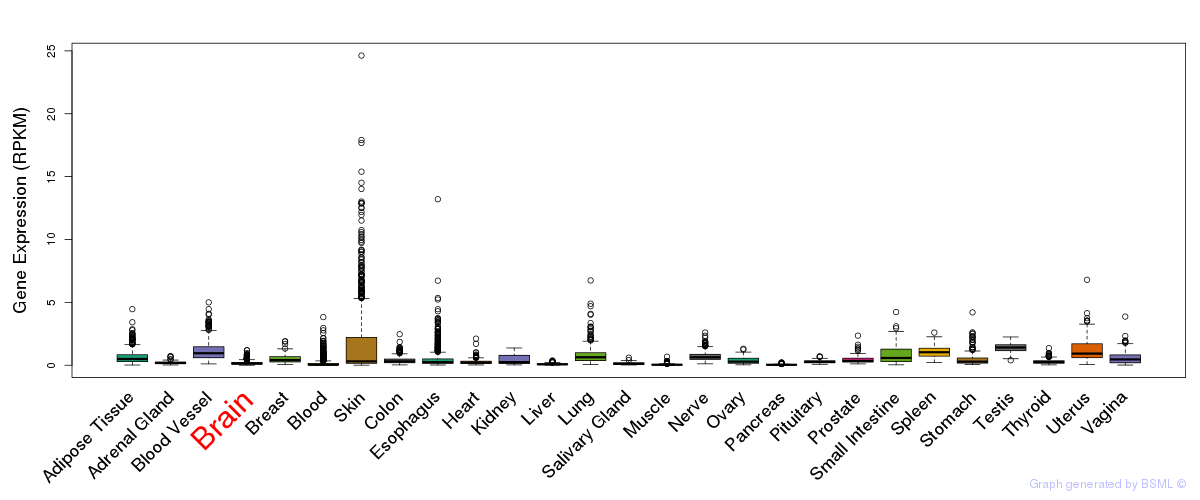
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KIAA0101 | 0.97 | 0.73 |
| CCNB2 | 0.97 | 0.80 |
| BIRC5 | 0.96 | 0.81 |
| CCNA2 | 0.96 | 0.76 |
| RRM2 | 0.96 | 0.80 |
| RAD51 | 0.96 | 0.72 |
| BUB1 | 0.95 | 0.81 |
| PBK | 0.95 | 0.76 |
| MELK | 0.95 | 0.79 |
| NEIL3 | 0.95 | 0.74 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FBXO2 | -0.43 | -0.67 |
| HLA-F | -0.43 | -0.69 |
| C5orf53 | -0.42 | -0.67 |
| PTH1R | -0.41 | -0.65 |
| ASPHD1 | -0.41 | -0.57 |
| SLC9A3R2 | -0.41 | -0.51 |
| AIFM3 | -0.41 | -0.67 |
| TINAGL1 | -0.40 | -0.68 |
| ALDOC | -0.40 | -0.65 |
| AF347015.27 | -0.40 | -0.73 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| HOOI ST7 TARGETS UP | 94 | 57 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 TARGETS UP | 457 | 269 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC1 AND HDAC2 TARGETS UP | 238 | 144 | All SZGR 2.0 genes in this pathway |
| SENESE HDAC2 TARGETS UP | 114 | 66 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| STONER ESOPHAGEAL CARCINOGENESIS UP | 39 | 25 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P7 | 90 | 52 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| OUILLETTE CLL 13Q14 DELETION DN | 60 | 38 | All SZGR 2.0 genes in this pathway |
| LE SKI TARGETS UP | 17 | 12 | All SZGR 2.0 genes in this pathway |
| KRISHNAN FURIN TARGETS DN | 13 | 8 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC ICP WITH H3K4ME3 | 445 | 257 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING | 243 | 155 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM B | 517 | 302 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
| NABA ECM REGULATORS | 238 | 125 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |